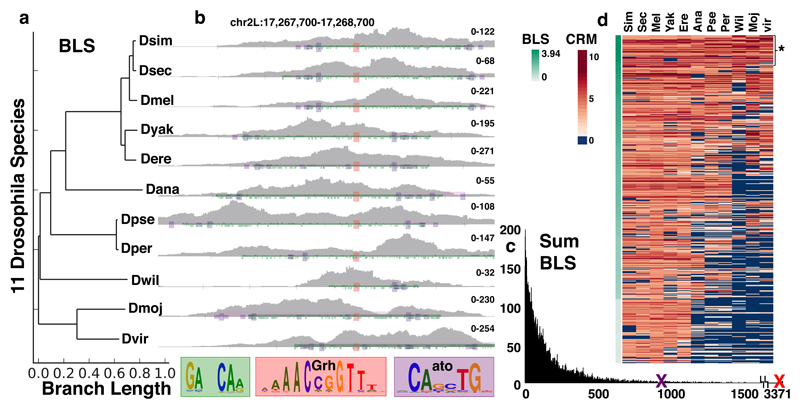
Figure 5. Identification of co-regulator motifs through evolutionary conservation.
(a) Branch Length Scores for the 11 Drosophila species. (b) ATAC signal from eye-antennal discs of 11 Drosophila species on a conserved Grh-Atonal enhancer (two eye-antennal disc ATAC-seq replicates per specie). The Grh motifs are shown in red, Atonal in purple and repeats in green. (c) Histogram of the cumulative Branch Length Scores for the 18k motifs. The top Grh motif (red X) has a score of 3371, the top Atonal motif (purple X) has a score of 878. (d) Heatmap showing the CRM scores for the Atonal motif across the conserved Grh enhancers ordered by BLS score. The top 480 enhancers are shown, with the 92 most conserved Grh-Ato enhancers (BLS > 3) marked by *.