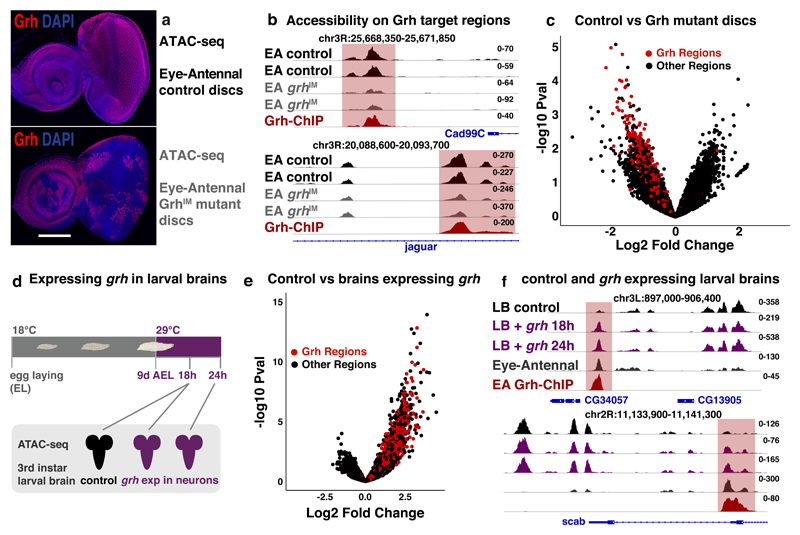
Figure 6. The pioneering role of Grh.
(a) Confocal images of control (top) and discs largely mutant for Grh (bottom), Grh C-terminal antibody68 staining (red), nuclei DAPI (blue) (reproducible results for 5 discs, scale bar = 100μm). (b) Tracks visualizing ATAC of control (black) and Grh mutant (grey) eye-antennal disc and Grh-ChIP (dark-red) in eye-antennal disc (2 biological replicates each). Grh target regions near epithelial genes Cad99C and jar lose accessibility. (c) Volcano plot showing the change in accessibility (DESeq2 log2FC and –log10 pval) of the 30774 regions in control versus Grh mutant eye-antennal discs, red dots mark the 3246 Grh regions. (d) Ectopic grh expression in the neurons of developing larvae. (e) Volcano plot showing the change in accessibility (DESeq2 log2FC and –log10 pval) of 37668 accessible regions in control larval brains versus larval brain with ectopic grh expression, red dots mark the 3246 Grh regions. (f) Tracks visualizing ATAC on control larval brains (black), larval brains with 18 and 24h grh expression (purple) (ATAC-seq experiments were done once for each time point) and eye-antennal discs (dark grey, 30 biological replicates), and Grh-ChIP on eye-antennal discs (dark-red, 3 biological replicates). Genomic loci near CG34057 and scab are shown with Grh target region (red).