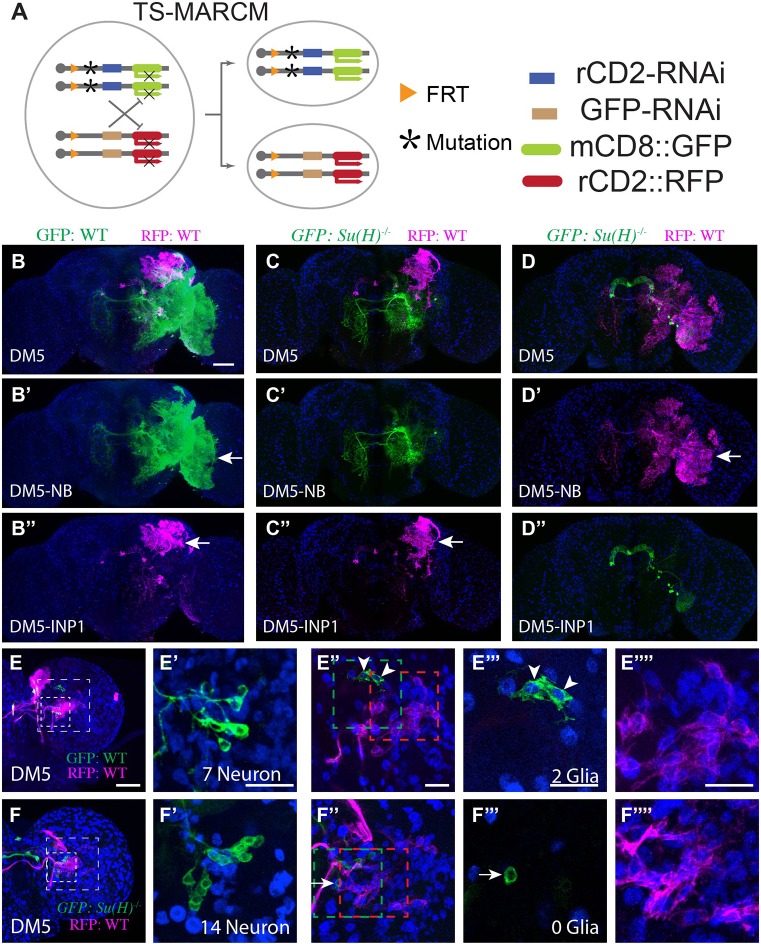
Fig. 5.
Notch is required for gliogenic switch. (A) Illustration of twin-spot MARCM, which marks the homozygous mutant clone with mCD8::GFP and its paired wild-type sister clone with rCD2::RFP. (B-D″) Su(H) mutant clones lack glia. Twin-spot MARCM clones of the DM5 lineage induced shortly after larval hatching and examined in adult fly brains counterstained with anti-Repo Ab (blue). (B-D) Merged views of B′-D′ and B″-D″. The glia-like processes (arrow) are present on both green and magenta sides of the control DM5 twin-spot clones (B-B″, n=3). By contrast, the glia-like processes are not seen on the mutant green side of the twin-spot MARCM clones derived from Su(H) heterozygous precursors (C′,D″, n=3 and 2, respectively). (E-F″″) Su(H) mutant clones have excess neurons at the expense of glia. Composite confocal images of the late larval brain carrying twin-spot MARCM clones. Partial projection of the INP cell body region (small box in E,F) and the glia region (big box in E,F) are shown in E′,F′ and E″,F″, respectively. Single confocal sections of INP glia (green box in E″,F″) and NB glia (E″″,F″; red box in E″,F″) are shown in E‴,F‴ and E″″,F″″, respectively. (E-E″″) Wild-type DM5 and DM5-INP1 twin-spot clone. The INP1 example shown has seven neurons (E′) and two glial cells (E‴). On average, DM5-INP1 has 7.7±0.5 neurons and 1.1±0.4 glial cells (n=7). Arrowheads indicate glial cells. (F-F″″) Wild-type DM5 and mutant DM5-INP1 twin-spot clone. The INP1 example shown has 14 neurons (F′) and no glia (F‴). The cell in F‴ is negative for Repo (arrow). On average, mutant INP1 has 13±2 neurons and 0±0 glial cells (n=4). Scale bars: 50μm in B-D″; 20 μm in E-F″″. Data are mean±s.d.