Fig. 1.
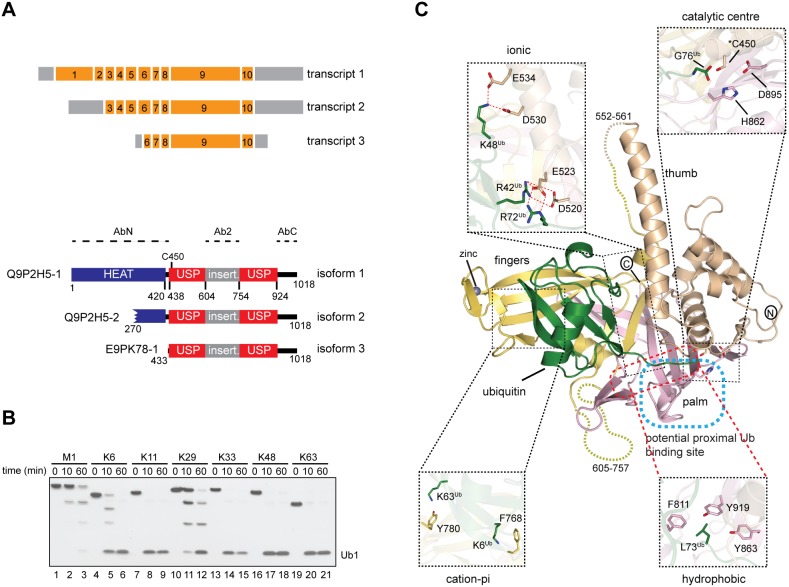
USP35 is an active and promiscuous DUB. (A) Upper panel, organisation of USP35 transcripts encoding isoforms presented in this study. Orange and grey boxes represent exons and non-coding regions, respectively. Exon numbers are given. Lower panel, domain organisation of USP35 isoforms. The USP domain of USP35 is shown together with catalytic Cys450 and an unstructured insertion spanning residues 604–753. The HEAT repeat motif is predicted based on the crystal structure of the N-terminus of a related protein, USP38 (PDB: 4RXX). Protein regions used to generate anti-USP35 antibodies are indicated. Uniprot accession numbers of each isoform are shown on the left. (B) Purified recombinant full-length human USP35 (375 nM) was incubated with tetraubiquitins of defined linkage types (733 nM) at 37°C for the indicated times. Reactions were resolved by SDS-PAGE followed by silver staining of the gel. (C) Crystal structure of the fragment of human USP35 (residues 423–944) lacking the insertion region (residues 604–753) and complexed with ubiquitin is presented. The subdomains thumb, palm and fingers are shown in beige, light pink and yellow, respectively, whereas the non-covalently bound distal ubiquitin is in green. The dashed line has been used to show the disordered regions. The co-ordinated zinc can be seen as a black sphere in the finger subdomain. A potential binding site of proximal ubiquitin is indicated. Close up views show the catalytic centre and selected residues mediating USP35–ubiquitin interactions.