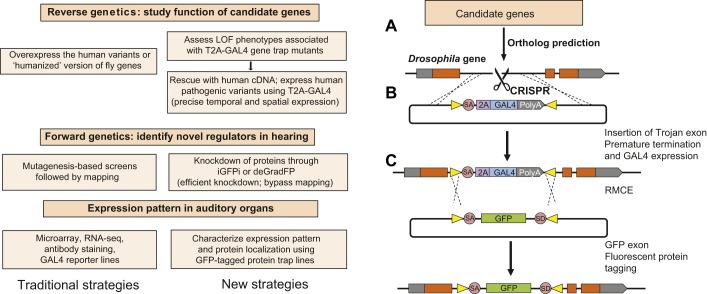
Fig. 5.
Approaches to functionally test candidate mammalian deafness genes in Drosophila. (A) Candidate genes can include known deafness genes or mouse genes with deafness phenotypes. Their Drosophila homologs are predicted by protein sequence homology. (B) An artificial exon that acts as a gene trap can be inserted into a MiMIC line by recombination-mediated cassette exchange (RMCE; Venken et al., 2011) or by CRISPR-mediated homology-directed repair, which prematurely terminates transcription of the endogenous gene through a splice acceptor site (SA) and poly-adenylation signal (PolyA), and expresses the GAL4 transcriptional activator as part of the endogenous transcript through a picornavirus self-cleaving 2A peptide sequence (2A). (C) Gene trap/GAL4 lines are screened for hearing phenotypes. The GAL4 protein expressed by the endogenous regulatory elements of the gene of interest can also be used to drive the expression of transgenes under the control of the UAS DNA element inserted elsewhere in the fly genome. These transgenes can be wild type, or pathogenic human or mouse orthologs, and the insertion of GAL4 into the endogenous locus allows the transgene to be expressed in a spatiotemporal pattern that is identical to the endogenous Drosophila gene. RMCE through attP and attB sites (yellow triangles) can convert the Trojan exon into an exon containing GFP flanked by SA and splice donor (SD) sites, to reveal the cellular and subcellular localization of the trapped gene (Nagarkar-Jaiswal et al., 2015b). The GFP exon can also be used to knockdown the encoded transcript or protein by in vivo GFP interference (iGFPi) or the deGradFP system, respectively. LOF, loss of function; RNA-seq, RNA sequencing.