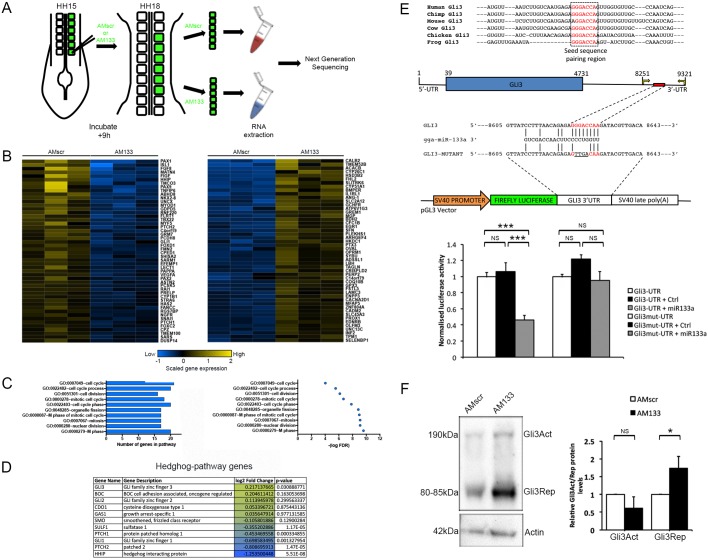
Fig. 3.
Differential transcriptomics reveals misregulation of Shh pathway components and identifies Gli3 as a direct miR-133 target. (A) Schematic overview of the experimental approach. Posterior somites of HH14/15 embryos were injected with FITC-labelled antagomir-133 (AM133) or scrambled-antagomir (AMscr) and harvested after 9 h for RNA isolation and sequencing. (B) Heatmaps of the top 50 genes significantly downregulated or upregulated after miR-133 KD shows clustering of six samples injected with AMscr or with AM133. (C) GO analysis showed that amongst the downregulated DE group, genes associated with cell cycle processes were significantly over-represented. The false discovery rates (FDRs) are shown for these genes. (D) Table showing Hedgehog (Hh) pathway genes that were de-repressed (yellow) and repressed (blue), including the transcriptional regulator Gli3, with log2 fold change and P-values. (E) Conservation of miR-133 seed sequence pairing region within the Gli3 3′UTR of different species. Luciferase assays validate Gli3 as a direct target for miR-133. Schematic of the chick Gli3 gene with a predicted target site containing an 8-mer seed match (red) present in the 3′UTR. Mutations (underlined) introduced into the predicted target site were designed to disrupt base pairing with the miR-133 seed region. A modified pGL3 vector containing a 1070 bp fragment of the chick Gli3 3′UTR downstream of the firefly luciferase reporter gene. Transfection of reporter plasmids into DF1 cells either on their own (white), or with a control miRNA mimic (black), or with miR-133 mimic (grey) confirms negative regulation of the reporter. The response was rescued after mutation of the target site. ***P<0.001; NS, not significant. (F) Western blot of somites injected with AMscr or AM133 shows increased amount of Gli3Rep protein. Quantitative analysis was performed on three biological replicates. *P<0.1; NS, not significant.