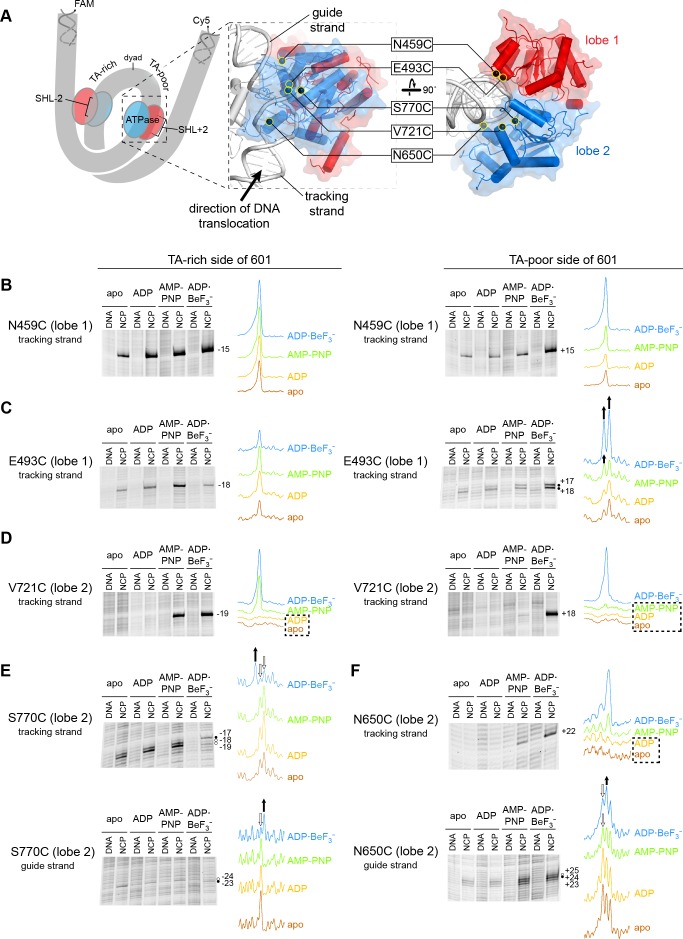
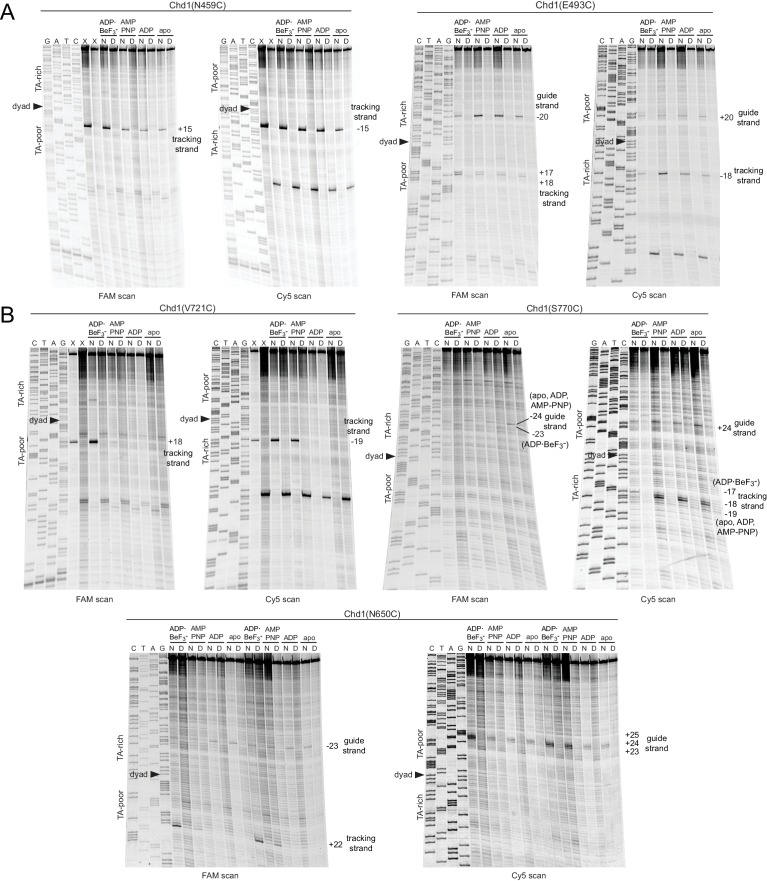
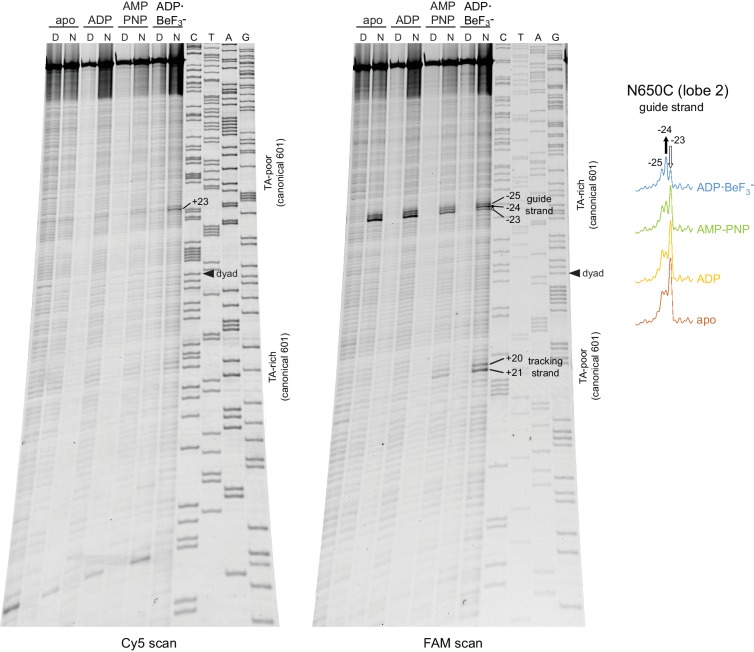
Figure 1. Interactions between the ATPase motor of Chd1 and nucleosomal DNA are sensitive to both nucleotide state and DNA sequence.
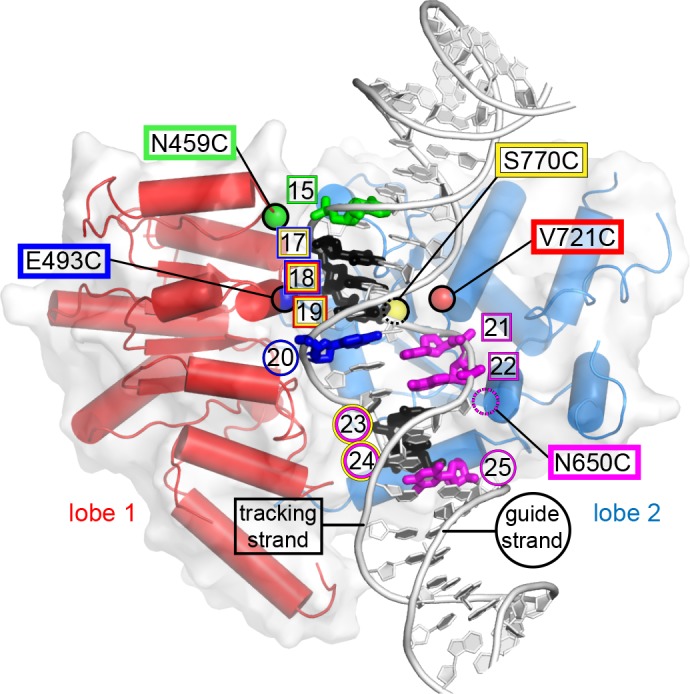
(A) Overview of the Chd1 ATPase motor on the nucleosome. A cartoon schematic (left) shows the superhelical wrap of nucleosomal DNA, with an ATPase motor (blue/red) at each of the superhelix location (SHL) 2 sites on either side of the nucleosome. The expanded views on the right show the molecular surface of the ATPase motor in complex with nucleosomal DNA, based on a cryoEM complex (Farnung et al., 2017). The five positions individually substituted with cysteine for APB cross-linking are highlighted with yellow circles, and the tracking and guide strands of DNA are indicated. The PyMOL visualization package (RRID:SCR_000305) was used for molecular rendering. (B-F). Representative data showing cross-linking to nucleosomal DNA on either the TA-rich side (left) or TA-poor side (right) of the 601 sequence, in different nucleotide conditions. Identical cross-linking reactions were performed with 300-450 nM Chd1 variant with either 150 nM canonical 601 nucleosomes with 40 bp of flanking DNA on each side ('NCP') or 150 nM naked DNA ('DNA'), to ensure observed cross-links were nucleosome-specific. Bands that increase or decrease in cross-linking efficiency for ADP·BeF3– compared to other conditions are indicated with black or white circles, respectively. Scans of each lane are shown as intensity plots to the right of each gel, with arrows highlighting bands that showed relative increases (black) or decreases (white) for ADP·BeF3– conditions. Dotted boxes highlight nucleotide conditions where cross-links were severely reduced or not detected. Numbering refers to the distances (nt) of cross-linked sites from the 601 dyad (positive numbers for TA-poor side and negative numbers for TA-rich side). These data are representative of 4 or more experiments. Extended gel images are shown in Figure 1—figure supplement 3.
Figure 1—figure supplement 1. Site-specific cross-linking visualized on the Chd1-nucleosome structure.