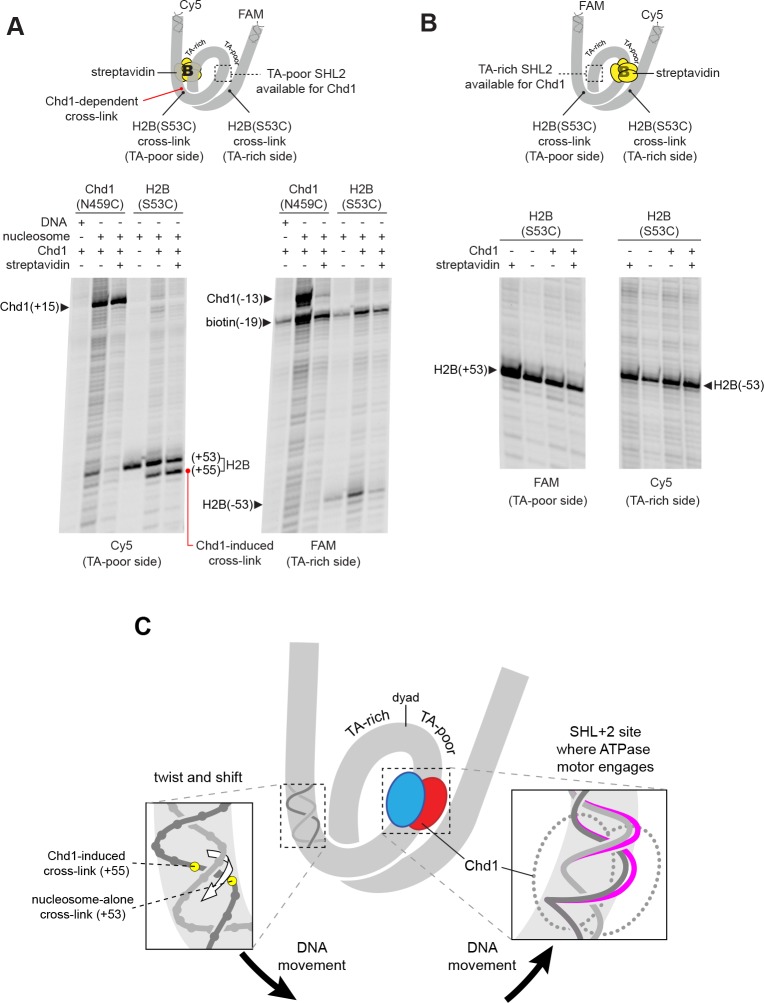
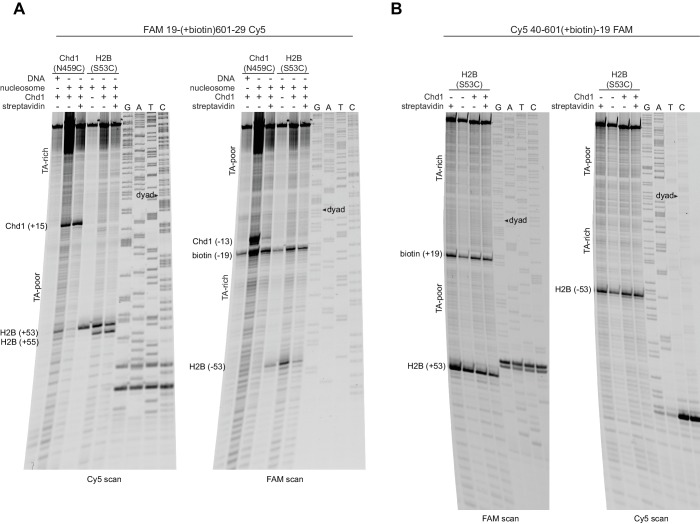
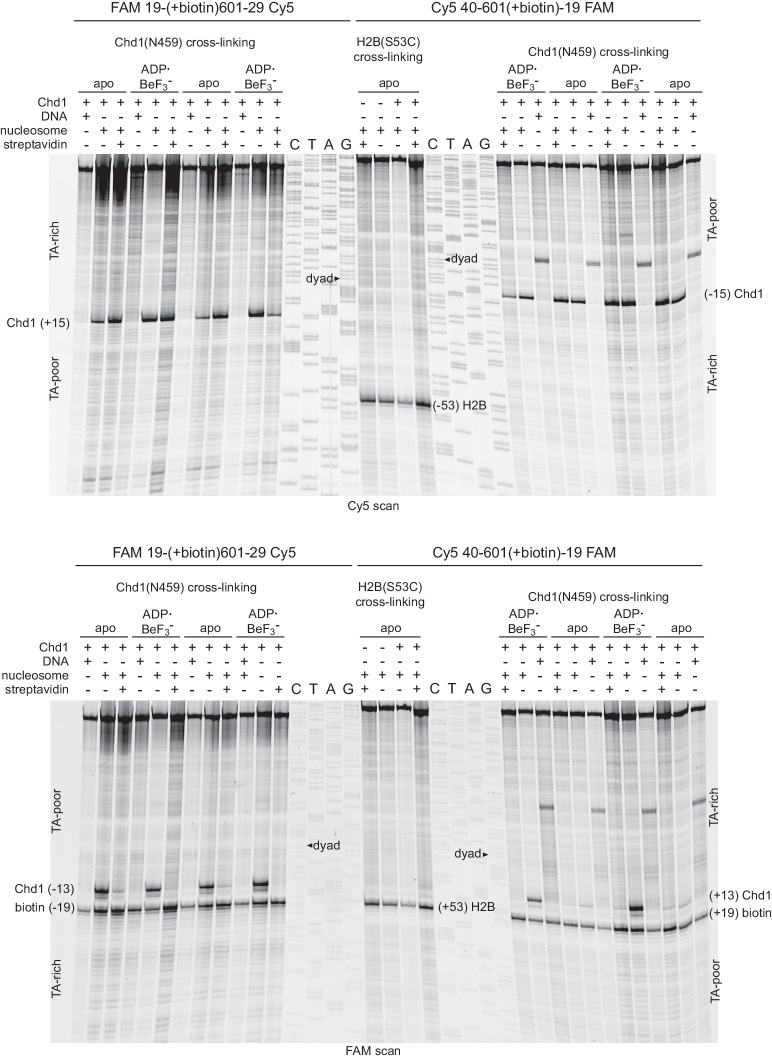
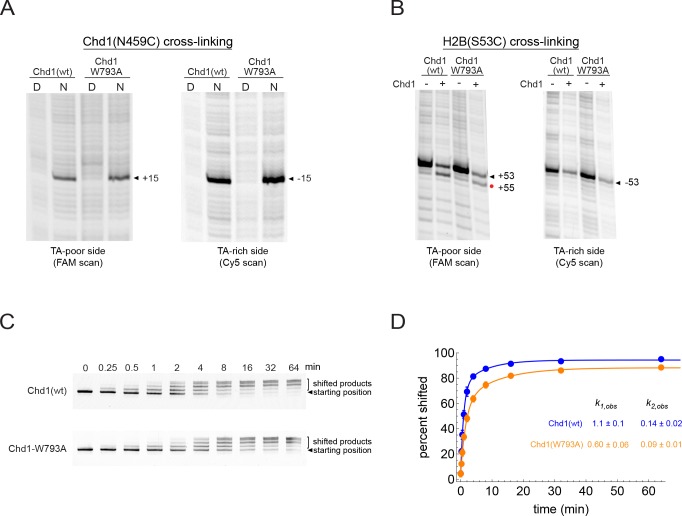
Figure 3. The Chd1-dependent shift in histone-DNA cross-linking correlates with Chd1 binding to the adjacent SHL2, on the same DNA gyre of the nucleosome.
(A) The presence of a biotin/streptavidin block at the TA-rich SHL–2 site does not diminish the Chd1-dependent shift in H2B(S53C) cross-links on the TA-poor side. The nucleosome substrate for these experiments was 19-[+biotin]601-29, with a biotin moiety 19 nt from the dyad on the TA-rich SHL–2 site. Note that for this nucleosome, the FAM and Cy5 labels are on opposite sides compared to all other 601 nucleosomes used in this study. Chd1(N459C) cross-linking experiments monitored Chd1 binding at each SHL2, and H2B(S53C) experiments followed the Chd1-dependent shift in histone-DNA cross-links on the TA-poor side. The apparent shift in Chd1(N459C) cross-linking site on the biotinylated strand (labeled as −13) was due in part to slower migration caused by the biotin moiety on the DNA fragment, and may not represent a true change in cross-linking position. The gels are representative of three or more experiments, and extended gel images are shown in Figure 3—figure supplement 1A. (B) The presence of a biotin moiety at the TA-poor SHL+2 site prevents the Chd1-dependent shift in H2B(S53C) cross-linking on the TA-poor side. The nucleosome substrate for these experiments was 40-601[+biotin]-19, with a biotin moiety 19 nt from the dyad on the TA-poor SHL+2 site. The absence of the H2B(S53C) shift likely stems from poor binding due to the biotin, as shown in Figure 3—figure supplement 2. The gels are representative of three or more experiments, and extended gel images are shown in Figure 3—figure supplement 1B. (C) A model for how Chd1 binding at SHL+2 may be coupled to shifts in DNA cross-linking.