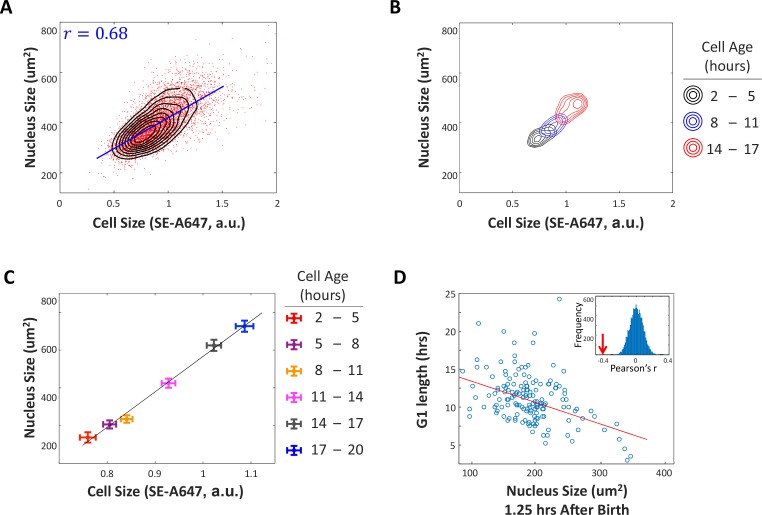
Figure 2. G1 length is negatively correlated with nuclear size at birth.
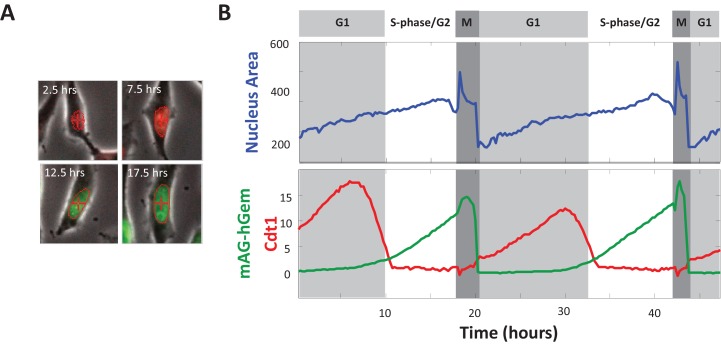
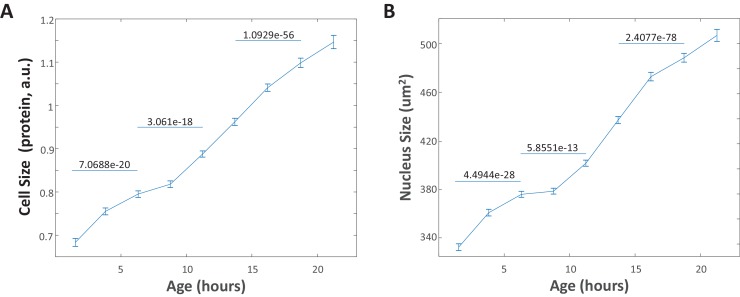
(A) A significant correlation between cell size and nucleus size suggests that nucleus size is an adequate proxy measurement of cell size. We imaged unsynchronized, unperturbed HeLa cells with time-lapse microscopy and monitored the size of their nuclei (estimated as the area covered by the nucleus in a widefield image), as a proxy for cell size. Datapoints (red) represent single-cell measurements of cell size (SE-A647 fluorescence) and nucleus size (projected area) in a population of HeLa cells. Also shown is the linear regression (blue line) between cell size and the size of the nucleus and the calculated Pearson correlation (r=0.68). Black contour lines represent the calculated joint probability density function, describing the frequency of cells for every given paired value of cell size and nucleus size. (B) Joint probability density functions of cell size and nucleus size for cells of different age groups, that is cells 2-5 hours after their most recent cell division (black), cells 8-11 hours after cell division (blue), and cells 14-17 hours after cell division (red). For (A–C), single cell measurements of cell age, cell size and nucleus size were performed on a large population of HeLa cells, as described for Figure 1. Measurements were performed in multiplex, such that all three parameters were quantified for each individual cell. We then sorted the single cell measurements into bins defined by cell age and separately calculated the joint probability of cell size and nucleus size for each age group. The figure demonstrates that the correlation of cell size and nucleus size consistently persists for cells of different ages. (C) Average cell size and nucleus size for cells of different age groups, as described in (B). This figure shows that as cells age, they increase in both nucleus size and cell size. Error bars represent standard error calculated as . The significant separation of the datapoints (in relation to errorbars) demonstrates that our cell size measurements and nucleus size measurements are sufficiently accurate to resolve average size differences that result from < 3 hours of growth, approximately 15% of cell cycle duration. (D) Size (area covered in widefield fluorescence image) of HeLa nucleus 1.25 hours after birth vs. G1 duration. Line shows least-squares linear fit. Pearson’s r = −0.41, p=6.7 x 10−8 (p-value calculated using a Student's t distribution for transformation of the correlation). Inset shows distribution of Pearson’s r-values generated by randomizing the data, with arrow marking the r-value of non-randomized data. Data shown in (D) are representative example of four biological replicates, n=158 cells. The raw data and source code necessary to generate (A–D) are included in Figure 2—source data 1.