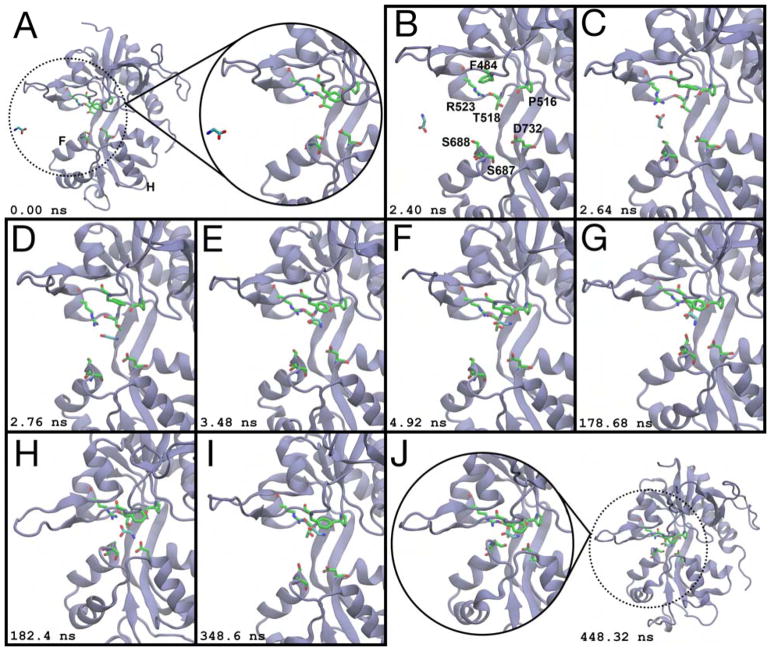
Figure 3.
Glycine binding to the GluN1 LBD. (A) Prior to ligand binding, glycine diffuses in bulk solvent, and the LBD is open. Helices F and H are labeled. (B) Glycine enters via the ξ1 side of the binding cleft. Binding pocket residues are labeled. (C) R523 contacts the glycine carboxylate. (D) Glycine moves into the binding pocket. (E) F484 interacts with the ligand’s amine. (F) The ligand rotates such that its amine contacts the T518 sidechain carboxylate and the P516 carbonyl oxygen. (G) The glycine amine rotates out of the binding pocket to interact with the S688 sidechain. (H) The amine contacts the sidechain carboxylate of D732, partially closing the LBD to (ξ1, ξ2) = (10.8 Å, 12.7 Å). (I) The LBD reopens, and glycine makes Lobe 1 contacts prior to cleft closure. (J) The LBD closes around the ligand to (ξ1, ξ2) = (9.2 Å, 10.3 Å). Glycine is unable to dissociate from this conformation. Snapshots of the binding process were taken from a 448 ns interval. Simulation time is given relative to panel (A). See also Movies S2 and S3.