Figure 2.
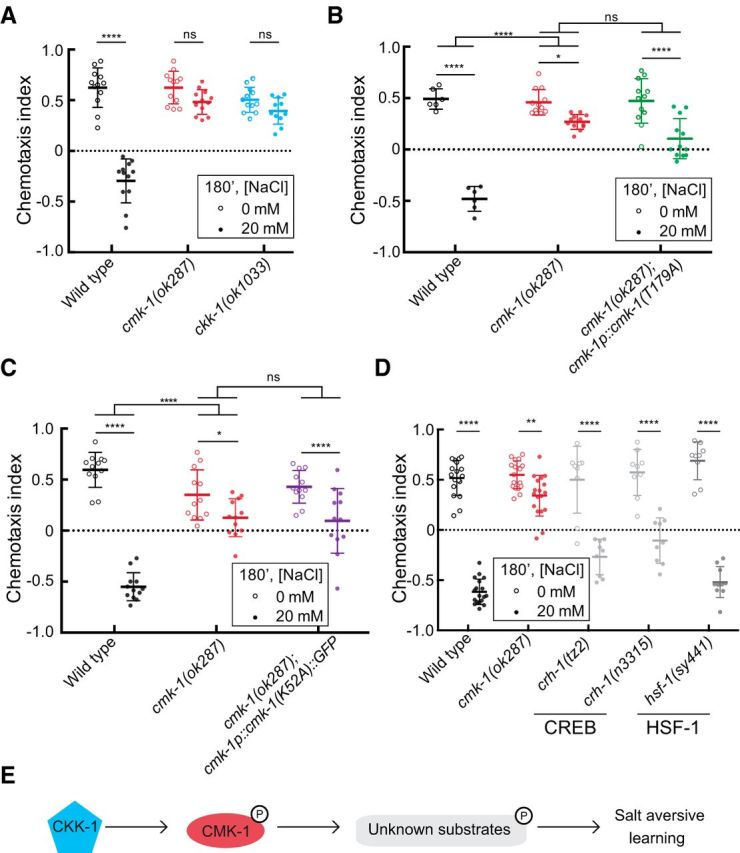
Salt-aversive learning requires CaMKK, CKK-1, and CMK-1 kinase activity. A, CKK-1 is needed for salt-aversive learning. A two-way ANOVA revealed a significant interaction between genotype and condition (F(2,65) = 47.37, p < 0.0001) and Sidak's post hoc test was used to test the effect of conditioning. B, A CMK-1 mutant that cannot be phosphorylated fails to restore salt-aversive learning to cmk-1 mutants. A two-way ANOVA across all genotypes and conditions revealed a significant interaction between genotype and condition (F(2,54) = 26.17, p < 0.0001) and Sidak's post hoc test revealed a significant effect of conditioning for all genotypes. A two-way ANOVA between wild-type and cmk-1(ok287) revealed a significant interaction between genotype and condition (F(1,32) = 112, p < 0.0001) and a two-way ANOVA between cmk-1(ok287) and cmk-1(ok287);cmk-1p::cmk-1(T179A) revealed a nonsignificant interaction between genotype and condition (F(1,44) = 3.521, p = 0.0672). C, A kinase-defective CMK-1(K52A) mutant protein cannot restore salt-aversive learning to cmk-1(ok287) mutants. A two-way ANOVA across all genotypes and conditions revealed a significant interaction between genotype and condition (F(2,66) = 33.96, p < 0.0001) and a Sidak's post hoc test revealed a significant effect of conditioning for all genotypes. A two-way ANOVA between wild-type and cmk-1(ok287) revealed a significant interaction between genotype and condition (F(1,44) = 71.16, p < 0.0001) and a two-way ANOVA between cmk-1(ok287) and cmk-1(ok287);cmk-1p::cmk-1(K52A)::GFP revealed a nonsignificant interaction between genotype and condition (F(1,44) = 0.6384, p = 0.4286). D, Behavior of known CMK-1 substrates. A two-way ANOVA revealed a significant interaction between genotype and condition (F(4,117) = 34.05, p < 0.0001). Sidak's post hoc test was used to assess the effect of conditioning for each genotype. For each panel in this figure, data were pooled from at least two independent experiments, consisting of three to six assays/condition performed blind to genotype. Animals of the indicated genotypes were conditioned for 180 min in either 0 or 20 mm NaCl and tested in a salt chemotaxis assay. Each symbol represents the results of a single assay; open and closed symbols indicate animals conditioned in 0 and 20 mm NaCl, respectively, and bars indicate mean ± SD of all assays conducted under the indicated condition and genotype. E, A schematic model, where CMK-1 is activated via phosphorylation by its upstream kinase CKK-1, and subsequently activates unknown downstream substrates to regulate salt-aversive learning. Sidak's posthoc tests were used to assess the effect of conditioning in panels A–D. The results are indicated by asterisks: *, **, and **** denote p < 0.1, p < 0.01, and p < 0.0001 and ns denotes not significant.
