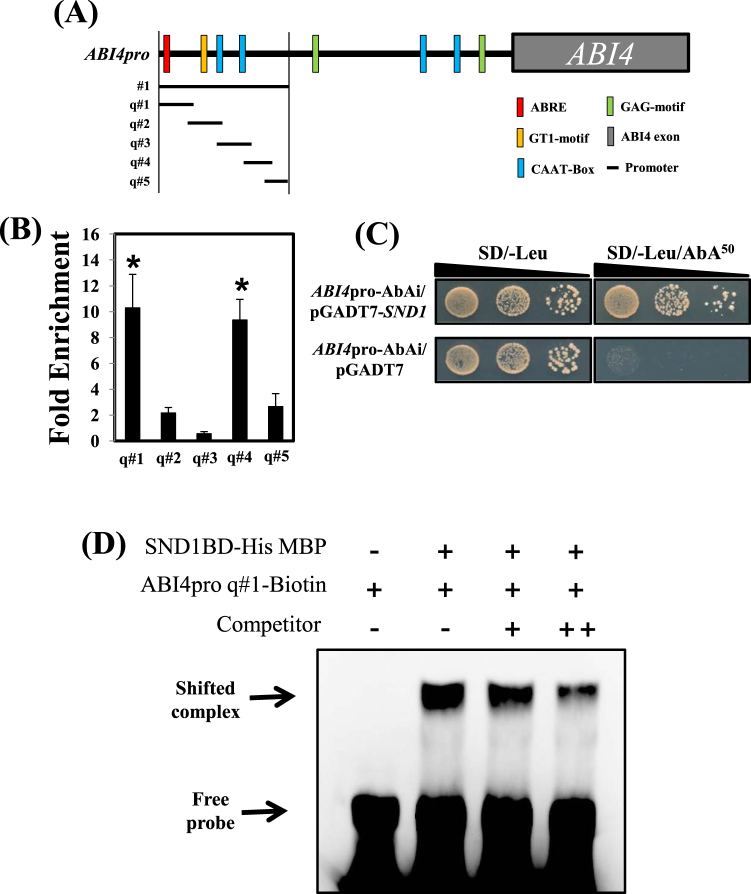
Figure 5.
Structure of the ABI4 promoter and chromatin immunoprecipitation (ChIP) using a SND1-GFP fusion protein. (A) cis-elements in the ABI4 promoter region. ABRE is an abscisic acid (ABA)-response element; GT1-motif and GAG-motif are light-response elements; CAAT-Box is a common cis-acting element. The PlantCARE program was used for ABI4 promoter analysis (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)19. ABI4pro#1, #1–1, and #1–2 show the size and location of the expected PCR products from the DNA template of ChIP. ABI4pro q#1, q#2, q#3, q#4, and q#5 present the size and location of the expected products of qRT-PCR products from the DNA template of ChIP. (B) The fold-enrichment values were calculated by dividing the ChIP signals obtained via the qRT-PCR by the signals obtained for the mock. The error bars indicate the standard error (SE). The asterisks represent significant differences among values (P < 0.05). (C) The yeast one-hybrid assay between SND1 and ABI4 promoter. Full-length SND1 was fused with GAL4 AD, and ABI4pro#1 was used as a bait. The ABI4pro-AbAi/pGADT7 yeast served as the negative control. Yeasts were cultured on SD/-Leu or SD/-Leu with 50 ng/mL aureobasidin A (AbA). (D) The electrophoretic mobility shift assay (EMSA) showed that the SND1 NAC domain (SND1 BD-His MBP) can bind to the biotin-labelled ABI4 promoter q#1.