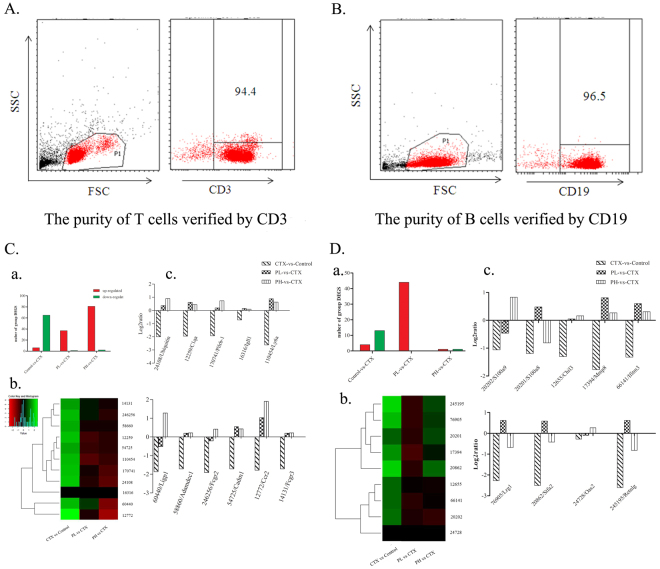
Figure 4.
Analysis of the polysaccharide formula on genes in T and B cells in immunosuppressed mice by DGE. (A,B) Lymphocytes from the spleen isolated by microbeads were stained with CD3-PE, and B220-PE (B cells) to test the purity. The figure shows that the isolated cells are pure. (C,a) Quantitative statistics of differentially expressed genes in B cells, the red shows up-regulated genes, and the green shows down-regulated genes. PL is the low dose group of polysaccharides with 200 mg/kg/bw and PH is the high dose of polysaccharide with 400 mg/kg/bw. (b) Hierarchical clustering of differential gene expression in B cells. The clustered map accorded by the log2 of significant difference multiples between the groups. Each row represents a gene and each column represents the comparison between the two groups, the red shows up-regulated genes, and the green shows down-regulated genes, the deeper the color the higher the gene expression. The gene ID is shown on the right, and the enrichment and the function of the displayed genes are shown, with references of the polysaccharide effect on these genes. (c) Statistics on the log2 value of the gene difference multiple in B map. The negative value is down-regulated and the positive value is up-regulated expression, and the 0 indicates no change. The figure shows that the formula of polysaccharide could restore the expression of the following genes in B cells: 24108/Ubiquitin, 12259/C1qa, 170741/Pilrb-1, 16316/Igll1, 110454/Ly6a, 60440/Ligp1, 58860/Adamdec1, 246256/Fcgr2, 54725/Cadm1, 12772/Ccr2 and 14131/Fcgr3. (D,a) Quantitative statistics of differentially expressed genes in T cells. (b) Hierarchical clustering of differential gene expression in T cells. The clustered map accorded by the log2 of significant difference multiples between the groups. (c) The figure shows that the compound polysaccharide could restore the expression of the following genes in T cells: 20202/S100a9, 20201/S100a8, 12655/Chil3, 17394/Mmp8, 66141/Ifitm3, 76905/Lrg1, 20862/Stfa2, 24728/Oas2 and 245195/Retnlg.