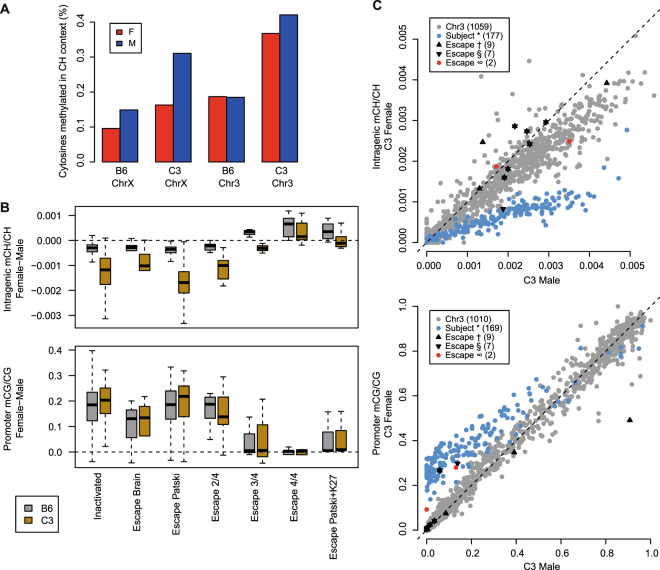
Figure 5.
Non-CpG methylation is enriched in genes escaping XCI in mouse liver. (A) The total fraction of methylated cytosines in CH (non-CpG) context by sex and strain for chromosomes X and 3 (Red, female; Blue, male). (B) Sex differences in intragenic mCH/CH and promoter mCG/CG distinguish escaper genes from inactivated genes. Boxplots of sex differences (female – male) in intragenic mCH/CH and promoter mCG/CG at genes with previously demonstrated XCI status in multiple mouse cell types. Classifications include genes subject to XCI in both brain and Patski cells (n = 177), brain-specific escapers (n = 6), Patski-specific escapers (n = 52), genes escaping in 2 of 4 (n = 9), 3 of 4 (n = 3), or 4 of 4 (n = 6) cell types analyzed by Berletch et al.42, or genes escaping in the Patski cell line and depleted of H3K27me3 in adult mouse liver (n = 7), as demonstrated by Yang et al.43. (C) Gene-level scatterplots comparing C3 male and C3 female intragenic mCH/CH and promoter mCG/CG for mouse genes by XCI status. Mouse genes reported as inactivated or escapers: Berletch et al. subject to XCI in both brain and Patski cells (*), Berletch et al. escape in at least 3 of 4 cell types (†), Yang et al. escape in Patski cells and depleted of H3K27me3 in adult liver (§). Predicted escaper genes (∞) and autosomal genes (Chr3) are also indicated.