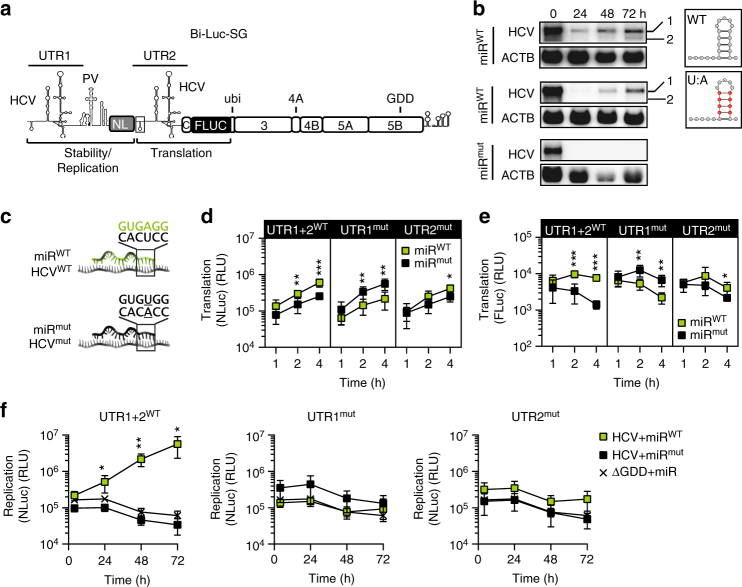
Fig. 2.
Dissection of stability and replication using a dual-luciferase replicon.a Illustration of the Bi-Luc-SG construct. miR-122 binding to UTR1 acts on RNA stability and replication (as measured by Nano luciferase activity), translation of non-structural proteins is driven by UTR2 (as measured by firefly luciferase activity). b Northern blot analysis of HCV RNA stability over 72 h. Stability of different Bi-Luc construct RNAs was analyzed in the presence of miRmut or miRWT. Band #1: full-length RNA, #2: truncated product. U:A mutations in SLI of UTR2 (red dots) prevented the formation of truncated RNA. c Illustration of miRWT (green) and miRmut (black) binding upon introduction of a matching point mutation in the target (underlined). d, e Translation assay of point mutants cloned into the MBR of UTR1 or 2. The activity of Nano (d) and firefly (e) luciferase was measured in the presence of miRmut or miRWT. f Assessment of the full intracellular replication cycle of the Bi-Luc-SG constructs after electroporation with miRmut or miRWT. Replication was monitored with the nano luciferase signal. Mean values (±SD), n = 3, duplicates. RLU relative light units, ΔGDD replication deficient mutant. For translation, statistical significance was determined for miRWT against miRmut, for replication miRWT and miRmut conditions were tested against ΔGDD. *P<0.05, **P<0.01, ***P<0.001