Abstract
Rickettsia felis, the causative agent of flea-borne spotted fever, occurs on all continents except Antarctica, owing to the cosmopolitan distribution of its cat flea vector. In this study, cat fleas were collected in two countries where the occurrence of R. felis was either unknown (Malta) or where accurate prevalence data were lacking (Israel). Altogether 129 fleas were molecularly analysed for the presence of rickettsial DNA. On the basis of three genetic markers, R. felis was identified in 39.5% (15/38) of the cat fleas from Malta. Sequences showed 100% identity to each other and to relevant sequences in GenBank. Among the 91 cat fleas from Israel, two (2.2%) contained the DNA of Candidatus Rickettsia senegalensis. Phylogenetically, the R. felis and Candidatus R. senegalensis identified here clustered separately (with high support) but within one clade, which was a sister group to that formed by the typhus group and spotted fever group rickettsiae. This is the first record of R. felis in Malta and of Candidatus R. senegalensis outside its formerly reported geographical range including Africa, Asia and North America.
Keywords: Emerging, gltA gene, ompA gene, phylogeny, Rickettsia, 17 kDa protein gene
Introduction
Rickettsiae are obligate intracellular Gram-negative bacteria which may affect their vertebrate hosts after arthropod-borne transmission [1]. Thus, the life cycle of pathogenic rickettsiae necessitates the presence of a blood-sucking vector [2]. The primary arthropod vectors and reservoirs vary according to Rickettsia spp., i.e. R. prowazekii and R. typhi in the typhus group (TG) are louse and flea borne [3], whereas nearly 20 Rickettsia species in the spotted fever group (SFG) are mite and tick borne [4]. In addition, R. felis, the causative agent of flea-borne spotted fever, is transmitted by the cat flea (Ctenocephalides felis), but it has also been demonstrated from a broad range of arthropods [2], [5].
During the last decade, R. felis–like organisms (RFLOs) have been identified with molecular methods in various arthropods, including cat fleas [5]. Among these, there are genetic variants, which (on the basis of their sequence divergence) have been proposed as new species, as exemplified by Rickettsia asembonensis [6] and Candidatus Rickettsia senegalensis [2]. The geographical distribution of RFLOs appears to be broad in a worldwide context, but their pathogenicity is unknown [7]. The sympatric occurrence of R. felis and RFLOs has also been reported [5], [8]. However, the role of RFLOs in modulating the vertical and horizontal transmission of other sympatric rickettsiae remains to be clarified [7].
Similar to RFLOs, R. felis occurs on all continents except Antarctica, owing to the cosmopolitan distribution of its vector, the cat flea [5]. During the past 15 years, approximately 30 countries were put on the map of flea-borne spotted fever [9], but there are regions without relevant information. The latter is exemplified by the middle and eastern regions of the Mediterranean Basin, where in several countries (including Malta) the occurrence of R. felis is unknown or actual/updated prevalence data are lacking (e.g. Israel; reported pool prevalence [10]). Therefore, in this study, cat fleas from Malta and Israel were molecularly analysed for the presence of rickettsial DNA.
Materials and methods
During the study, the following numbers of cat fleas were collected in 2017: 38 from 11 cats and three dogs at eight locations (data not shown) in Malta, and 91 specimens from 28 cats and three dogs in Jerusalem, Israel. Fleas were removed from these animals during regular veterinary care; therefore, no ethical permission was needed. Fleas from each host were stored in 96% ethanol separately, and their species was identified according to Whitaker [11].
DNA was extracted from individual fleas with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions, including an overnight digestion in tissue lysis buffer and proteinase K at 56°C. Extraction controls were used to monitor cross-contamination among samples. All samples were tested for the quantity and quality of DNA contents with a TaqMan real-time PCR specific for the 18S rRNA gene (Thermo Fisher Scientific, Vantaa, Finland) [12].
Flea DNA extracts were screened for the presence of rickettsiae with a TaqMan PCR amplifying a 74 bp fragment of the citrate synthase (gltA) gene of SFG and TG rickettsiae [12]. From the positive samples, a 796 bp long fragment of the gltA gene was amplified for sequencing with primers CS477f and CS1273r [13], as previously described [12]. In addition, an approximately 480 bp long fragment of the 17 kDa surface antigen gene of Rickettsia spp. was amplified with primers 17kd1 (5′-GCT CTT GCA ACT TCT ATG TT-3′) and 17kd2 (5′-CAT TGT TCG TCA GGT TGG CG-3′) [14]. The 25.0 μL final volume of reaction mixture contained 5.0 μL template DNA, 0.5 U HotStar Taq Plus DNA Polymerase (5 U/μL) (Qiagen), 2.5 μL of 10 × Coral Load PCR buffer (15 mM MgCl2 included), 0.5 μL dNTP Mix (10 mM), 0.5 μL of each primer (50 μM) and 15.9 μL distilled water. The thermal cycle included an initial denaturation step at 95°C for 5 minutes, followed by 40 cycles of denaturation at 95°C for 30 seconds, annealing at 51°C for 30 seconds and extension at 72°C for 1 minute. Final extension was performed at 72°C for 5 minutes. In a fourth PCR, an approximately 532 bp long fragment of the outer membrane protein A (ompA) gene of Rickettsia spp. was amplified with primers Rr190.70p (5′-ATG GCG AAT ATT TCT CCA AAA-3′) and Rr190.602n (5′-AGT GCA GCA TTC GCT CCC CCT-3′) [15]. Conditions were the same as above, except using 1.0 U polymerase and annealing at 48°C for 30 seconds.
PCR products of the 17 kDa and ompA genes were sequenced at Biomi (Gödöllő, Hungary) and those of the gltA gene at Microsynth (Balgach, Switzerland). Sequences were edited and assembled using Geneious 9.1.7 (http://www.geneious.com [16]), then aligned and compared to reference GenBank sequences by the nucleotide BLASTn programme (https://blast.ncbi.nlm.nih.gov). Representative sequences were submitted to GenBank (R. felis from Malta: MG893575 (gltA gene), MG893577 (17 kDa antigen gene), MG893579 (ompA gene); Candidatus R. senegalensis from Israel: MG893576 (gltA gene), MG893578 (17 kDa antigen gene)). Phylogenetic analyses were performed by the maximum-likelihood method and Tamura3 model using MEGA 6.0. Exact confidence intervals for the prevalence rates were calculated at the 95% level.
Results
Among the 38 DNA extracts of cat fleas from Malta, 15 (39.5%; 95% confidence interval, 24–56.6) were gltA PCR positive for rickettsiae. In all of these samples, R. felis was identified by sequencing, with 100% (757/757 bp) identity to each other and to R. felis sequences in GenBank (JQ674484 from Gabon, AF210692 from the United States). The amplified parts of the 17 kDa and ompA genes were also 100% (385/385 bp and 452/452 bp, respectively) identical with those of R. felis (e.g. KF241853 and AJ563398, respectively, from Mexico).
Among the 91 DNA extracts of cat fleas from Israel, two (2.2%; 95% confidence interval, 0.3–7.7) were gltA PCR positive for rickettsiae. In both of these samples Candidatus R. senegalensis was identified by sequencing, with 100% identity to each other and to two conspecific sequences in GenBank (757/757 bp identity with KF666472 from Senegal, 736/736 bp identity with KU499847 from India). The amplified part of the 17 kDa gene was also 100% (385/385 and 375/375 bp) identical with that of Candidatus R. senegalensis reported from the United States (AY953285 and KU167051-2, respectively). Amplification of the ompA gene was not successful from these samples.
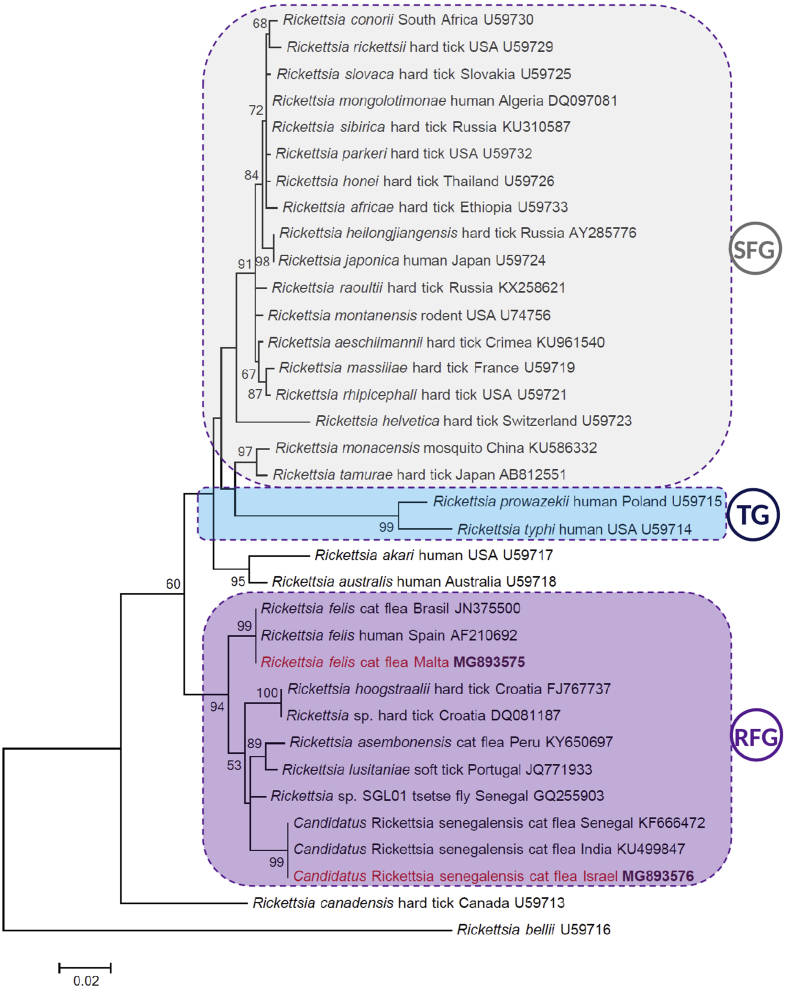
Phylogenetically, R. felis and Candidatus R. senegalensis identified here clustered separately (with 94% bootstrap support) but within one clade, which was a sister group to that formed by TG and SFG rickettsiae (Fig. 1).
Fig. 1.
Maximum-likelihood tree of Rickettsia spp. based on gltA sequences. Two sequences identified in this study are shown in red with bold accession numbers. Further sequences of Rickettsia spp. (representing major phylogenetic groups and having nearly 100% coverage with sequences obtained here) were retrieved from GenBank. Clades of RFG (Rickettsia felis group), SFG (spotted fever group) and TG (typhus group) are circled by dashed line filled with different colours. Species name, isolation source and country of origin are shown for each entry. Branch lengths represent number of substitutions per site, inferred according to scale shown.
Discussion
Epidemiologic studies of R. felis and related bacteria are considered to be important because their natural cycles have not yet been fully elucidated [2]. The recent emergence of R. felis–associated febrile diseases in West and East Africa further justifies this [2]. While clinical cases of R. felis infection in humans are well documented, the pathogenicity of Candidatus R. senegalensis is unknown. However, a Rickettsia species with similar sequence (with 98.5%, i.e. 673/683 bp, identity) has been reported in Senegal from a human patient with febrile illness (JQ674485) [2].
Assessing the significance of the present findings in a geographical context, the first record of R. felis in Malta provides new information on the country level and suggests a relatively high epidemiologic risk of human infection in that island. For comparison, 25.8% of cat fleas were shown to carry R. felis in Sicily, southern Italy [17], which is a lower prevalence rate compared to the 39.5% found here.
However, the first detection of Candidatus R. senegalensis in Israel is new on the continental level because this species has been hitherto reported at great distances from the Middle East, i.e. in Africa (Senegal, KF666472), Asia (India, KU499847) and North America (United States, AY953285, KU167051). In addition, a Rickettsia genotype which has highly similar gltA sequence to Candidatus R. senegalensis was identified in Asia (Thailand, AF516331) and a further one with a similar sequence in Africa (Ivory Coast, JN620082; Gabon, JQ354961). At the same time, the gltA sequence identified here in cat fleas from Israel was only 96% (328/340 bp) similar to an RFLO (under accession no. KP050777) formerly identified in desert fleas in Israel [18].
Phylogenetic analyses of R. felis and Candidatus R. senegalensis were performed in a previous study [2]. On the basis of the gltA gene, it was established that these two species, together with Rickettsia hoogstraalii, form a compact, isolated clade that is a sister group to the neighbouring Rickettsia akari and Rickettsia australis cluster [2]. However, in that phylogenetic analysis, SFG rickettsiae appeared to be underrepresented. By including a higher number of SFG Rickettsia spp. in the phylogenetic analysis here, not only was the separateness of the R. felis clade confirmed, but this also became a sister group to the cluster containing all SFG and TG species, as well as R. akari and R. australis (Fig. 1). This pattern was confirmed in maximum-likelihood analyses with other models (Hasegawa-Kishino-Yano and Tamura-Nei; tree not shown). Therefore, inclusion of the R. felis cluster in phylogenetic trees [2] argues against the formerly reported basal position of the TG among rickettsiae [1].
This pattern of phylogenetic clustering of R. felis and closely related species or genotypes was confirmed with other molecular markers [2] except ompA. Concerning the latter, it has been reported that the amplification of the ompA gene typical for the SFG of rickettsiae was unsuccessful in the case of Candidatus R. senegalensis [2], similar to what was experienced in this study.
In conclusion, the results of the present study broaden the geographical range of R. felis and Candidatus R. senegalensis. These findings also prompt extension of molecular analyses of cat fleas (the flea species with the most cosmopolitan distribution) for rickettsiae in those countries, where relevant data are lacking.
Acknowledgements
SH was supported by OTKA 115854. Part of the molecular work was performed using the logistics of the Center for Clinical Studies, Vetsuisse Faculty, Zurich, Switzerland.
Conflict of interest
None declared.
References
- 1.Sekeyova Z., Roux V., Raoult D. Phylogeny of Rickettsia spp. inferred by comparing sequences of ‘gene D,’ which encodes an intracytoplasmic protein. Int J Syst Evol Microbiol. 2001;51:1353–1360. doi: 10.1099/00207713-51-4-1353. [DOI] [PubMed] [Google Scholar]
- 2.Mediannikov O., Aubadie-Ladrix M., Raoult D. Candidatus ‘Rickettsia senegalensis’ in cat fleas in Senegal. New Microbes New Infect. 2015;3:24–28. doi: 10.1016/j.nmni.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillespie J.J., Ammerman N.C., Beier-Sexton M., Sobral B.S., Azad A.F. Louse- and flea-borne rickettsioses: biological and genomic analyses. Vet Res. 2009;40:12. doi: 10.1051/vetres:2008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood H., Artsob H. Spotted fever group rickettsiae: a brief review and a Canadian perspective. Zoonoses Public Health. 2012;59(Suppl. 2):65–79. doi: 10.1111/j.1863-2378.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown L.D., Macaluso K.R. Rickettsia felis, an emerging flea-borne rickettsiosis. Curr Trop Med Rep. 2016;3:27–39. doi: 10.1007/s40475-016-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maina A.N., Luce-Fedrow A., Omulo S., Hang J., Chan T.C., Ade F. Isolation and characterization of a novel Rickettsia species (Rickettsia asembonensis sp. nov.) obtained from cat fleas (Ctenocephalides felis) Int J Syst Evol Microbiol. 2016;66:4512–4517. doi: 10.1099/ijsem.0.001382. [DOI] [PubMed] [Google Scholar]
- 7.Blanton L.S., Walker D.H. Flea-borne rickettsioses and rickettsiae. Am J Trop Med Hyg. 2017;96:53–56. doi: 10.4269/ajtmh.16-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maina A.N., Fogarty C., Krueger L., Macaluso K.R., Odhiambo A., Nguyen K. Rickettsial infections among Ctenocephalides felis and host animals during a flea-borne rickettsioses outbreak in Orange County, California. PLoS One. 2016;11:e0160604. doi: 10.1371/journal.pone.0160604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legendre K.P., Macaluso K.R. Rickettsia felis: a review of transmission mechanisms of an emerging pathogen. Trop Med Infect Dis. 2017;2:64. doi: 10.3390/tropicalmed2040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer O., Baneth G., Eshkol T., Shaw S.E., Harrus S. Polygenic detection of Rickettsia felis in cat fleas (Ctenocephalides felis) from Israel. Am J Trop Med Hyg. 2006;74:444–448. [PubMed] [Google Scholar]
- 11.Whitaker A.P. vol. 1. Field Studies Council; Shrewsbury, UK: 2007. Fleas—siphonaptera. (Handbooks for the identification of British insects). [Google Scholar]
- 12.Boretti F.S., Perreten A., Meli M.L., Cattori V., Willi B., Wengi N. Molecular investigations of Rickettsia helvetica infection in dogs, foxes, humans, and Ixodes ticks. Appl Environ Microbiol. 2009;75:3230–3237. doi: 10.1128/AEM.00220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekeyova Z., Fournier P.E., Rehacek J., Raoult D. Characterization of a new spotted fever group rickettsia detected in Ixodes ricinus (Acari: Ixodidae) collected in Slovakia. Med Entomol. 2000;37:707–713. doi: 10.1093/jmedent/37.5.707. [DOI] [PubMed] [Google Scholar]
- 14.Williams S.G., Sacci J.B., Schriefer M.E., Anderson E.M., Fujioka K.K., Sorvillo F.J. Typhus and typhus-like rickettsiae associated with opossums and their fleas in Los Angeles county, California. J Clin Microbiol. 1992;30:1758–1762. doi: 10.1128/jcm.30.7.1758-1762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regnery R.L., Spruill C.L., Plikaytis B.D. Genotypic identification of rickettsiae and estimation of interspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. 1991;173:1576–1589. doi: 10.1128/jb.173.5.1576-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giudice E., Di Pietro S., Alaimo A., Blanda V., Lelli R., Francaviglia F. A molecular survey of Rickettsia felis in fleas from cats and dogs in Sicily (Southern Italy) PLoS One. 2014;9:e106820. doi: 10.1371/journal.pone.0106820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rzotkiewicz S., Gutiérrez R., Krasnov B.R., Morick D., Khokhlova I.S., Nachum-Biala Y. Novel evidence suggests that a ‘Rickettsia felis–like’ organism is an endosymbiont of the desert flea, Xenopsylla ramesis. Mol Ecol. 2015;24:1364–1373. doi: 10.1111/mec.13106. [DOI] [PubMed] [Google Scholar]