Figure 1.
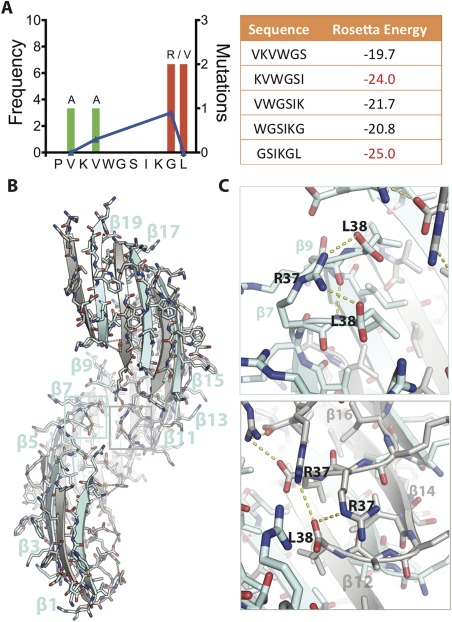
Crystal structure of SOD1 segment harboring a familial mutation, G37R. (A) (Left) Frequency of ALS‐associated mutations in SOD‐1 residues 28–38. Data from http://alsod.iop.kcl.ac.uk. (Right) Computed energies of steric zippers formed by six‐residue segments within segment 28–38 by ZipperDB.18 Segments KVWGSI and GSIKGL have energies of −23 kcal/mol or lower and are predicted to form fibrils. (B) 2.9 Å crystal structure of the toxic oligomeric core of SOD1, residues 28–38 with the familial mutation G37R, reveals a corkscrew assembly of antiparallel β‐strands with a hydrophobic cleft composed of valine and isoleucine residues. Twenty strands form one complete turn with a pitch length of 59 Å and diameter of 38 Å. (C) Arg37 forms multiple inter‐ and intra‐chain interactions that increase the stability of the assembly. (Top) Shown here Arg37 of β7 forms an intra‐chain hydrogen bond with Leu38 and an inter‐chain hydrogen bond with Leu38 of β9. (Bottom) Arg37 also forms inter‐chain hydrogen bonds. Shown here Arg37 of β14 is engaged in hydrogen bonds with Leu38 of β12 and Arg37 of β16. Together these interactions lead to 18 additional hydrogen bonds per turn.
