Abstract
Background
Glioblastoma (GBM) is the most common adult primary brain tumor. Multimodal treatment is empiric and prognosis remains poor. Recurrent PIK3CA missense mutations (PIK3CAmut) in GBM are restricted to three functional domains: adaptor binding (ABD), helical, and kinase. Defining how these mutations influence gliomagenesis and response to kinase inhibitors may aid in the clinical development of novel targeted therapies in biomarker-stratified patients.
Methods
We used normal human astrocytes immortalized via expression of hTERT, E6, and E7 (NHA). We selected two PIK3CAmut from each of 3 mutated domains and induced their expression in NHA with (NHARAS) and without mutant RAS using lentiviral vectors. We then examined the role of PIK3CAmut in gliomagenesis in vitro and in mice, as well as response to targeted PI3K (PI3Ki) and MEK (MEKi) inhibitors in vitro.
Results
PIK3CAmut, particularly helical and kinase domain mutations, potentiated proximal PI3K signaling and migration of NHA and NHARAS in vitro. Only kinase domain mutations promoted NHA colony formation, but both helical and kinase domain mutations promoted NHARAS tumorigenesis in vivo. PIK3CAmut status had minimal effects on PI3Ki and MEKi efficacy. However, PI3Ki/MEKi synergism was pronounced in NHA and NHARAS harboring ABD or helical mutations.
Conclusion
PIK3CAmut promoted differential gliomagenesis based on the mutated domain. While PIK3CAmut did not influence sensitivity to single agent PI3Ki, they did alter PI3Ki/MEKi synergism. Taken together, our results demonstrate that a subset of PIK3CAmut promote tumorigenesis and suggest that patients with helical domain mutations may be most sensitive to dual PI3Ki/MEKi treatment.
Introduction
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults [1]. It is also the most aggressive, with a median survival of only 12–15 months [1–3]. The molecular heterogeneity of GBM has been extensively characterized [4–6]. The vast majority of GBM arise de novo and harbor frequent mutations in 3 “core” signaling pathways: RB, TP53, and receptor tyrosine kinase (RTK)/mitogen activated protein kinase (MAPK)/phosphoinositide 3-kinase (PI3K) [5]. GBM can be stratified into 4 molecular subtypes based on gene expression [6]. However, this knowledge has yet to impact patient management. First line therapy remains empiric and consists of surgical resection followed by radiation with concurrent and adjuvant temozolomide, a DNA damaging agent [3]. Clinical trials of inhibitors targeting the pathways frequently mutated in GBM have had disappointing results for a variety of reasons, including drug resistance and inclusion of molecularly heterogeneous patients [7, 8]. Preclinical modeling can aid in development of novel therapies by defining whether mutations associated with GBM drive disease pathogenesis and are predictive of drug response.
The PI3K pathway promotes many cancer hallmarks, including survival, proliferation, and migration/invasion [9–12]. PI3K is a heterodimeric lipid kinase composed of catalytic and regulatory subunits encoded by genes such as PIK3CA and PIK3R1, respectively [13, 14]. Pathway activation is mediated by the phosphorylation of PIP2 to PIP3 by the catalytic subunit, resulting in recruitment and activation of effector proteins, including AKT. PI3K signaling (hereafter PI3K) is antagonized by the tumor suppressor PTEN [13, 14]. The PI3K pathway is an attractive therapeutic target in GBM because mutually exclusive mutations in PIK3CA, PIK3R1, and PTEN occur in 46% of patients [15–17].
PI3K activation via Pten deletion, PIK3R1 mutation, or constitutive AKT promotes tumorigenesis in multiple preclinical GBM models [18–23]. For example, we found that Pten deletion cooperates with mutant Kras to activate PI3K and potentiate malignant progression in immortalized mouse astrocytes [19, 24]. Similarly, constitutive AKT cooperated with mutant RAS to promote tumorigenesis in immortalized human astrocytes (NHA) [22, 25]. However, the role of PIK3CA mutations in gliomagenesis has not been experimentally investigated.
PIK3CA is altered in 10% of GBM, mostly via missense mutations [4, 15, 16]. These mutations are generally restricted to 3 functional protein domains (adaptor binding (ABD), helical, and kinase) and are predicted to activate PI3K via distinct biochemical mechanisms [26]. Some PIK3CAmut found in GBM have been shown to promote tumorigenesis in non-brain tissues, particularly the most prevalent helical (E542K, E545K) and kinase (H1047R) domain mutations [27–29]. However, their role in gliomagenesis has yet to be determined. Here we defined the role of PIK3CAmut in GBM pathogenesis using NHA and NHARAS. Furthermore, we determined whether these mutations influenced response to single agent and combination PI3K/MEK inhibitors buparlisib and selumetinib, respectively, to elucidate the utility of PIK3CAmut as a predictive biomarker.
Buparlisib (BKM120) has been proposed as a potential targeted therapy for GBM [30–33]. It is currently being investigated in a Phase II clinical trial in GBM patients (ClinicalTrials.gov, NCT01339052). While there have been no GBM clinical trials of selumetinib, we and others have shown its efficacy in preclinical models [34, 35]. Enriching future clinical trials with likely responders based on mutational profiles promises to improve the chances of clinical success.
Materials and methods
Supplement
Supplemental methods, figures, and tables can be found online.
PIK3CA mutagenesis and lentivirus production
Third generation lentiviral gateway destination vector (pLenti-PGK-Hygro-DEST, #19066, a gift from Eric Campeau and Paul Kaufman), pENTR4 vector (pENTR4-no-ccDB, #17424, a gift from Eric Campeau and Paul Kaufman), hemagglutinin (HA)-tagged wild-type (WT) PIK3CA (PIK3CAWT, pBabe-puro-HAPIK3CA, #12522, a gift from Jean Zhao), and HA-tagged GFP (GFP, pDEST-Flag-HA-GFP, #22612, a gift from Wade Harper) plasmids were purchased from Addgene (Cambridge, MA) [36–38]. Wild-type PIK3CA (PIK3CAWT) and GFP were excised and inserted into pENTR4 vector by ligation. PIK3CAmut (R88Q, C90Y, E542K, E545K, M1043V, H1047R) were generated by point mutagenesis of PIK3CAWT using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA) per manufacturer’s instructions. GFP, PIK3CAWT, and PIK3CAmut were transferred from pENTR4 to pLenti-PGK-Hygro-DEST vectors by recombination as described [36]. All mutations were confirmed by Sanger sequencing (Genewiz, South Plainfield, NJ). Lentiviral particles encoding GFP, PIK3CAWT, or individual PIK3CAmut were generated in 293FT cells (Invitrogen, Grand Island, NY) per manufacturer’s instructions.
Cell culture
NHA and NHARAS lines were a kind gift from Russell O. Pieper [25]. Cells were maintained as adherent cultures at 37°C and 5% CO2 in DMEM supplemented with 5% FBS and 1% penicillin/streptomycin (complete DMEM). To generate NHA and NHARAS lines expressing GFP, PIK3CAWT, or PIK3CAmut, 135,000 and 120,000 cells respectively were plated on 60 cm2 plates. Lentiviruses were added two days after plating, then incubated with cells overnight in complete DMEM containing 8 μg/ml polybrene (Sigma-Aldrich, St. Louis, MO) at 37°C and 5% CO2. Two days post-infection, transduced cells were selected by culture in complete DMEM plus 300 μg/ml hygromycin B (Gold Biotechnology, St. Louis, MO) for 14 days. Stable gene expression was confirmed by immunoblot for the HA tag on PIK3CAwt and PIK3CAmut. All in vitro experiments were performed in DMEM with 2.5% FBS and 1% penicillin/streptomycin (low serum medium) unless otherwise stated.
Drugs
The PI3K inhibitor (PI3Ki) buparlisib (BKM120) and the MEK inhibitor (MEKi) selumetinib (AZD6244) were purchased from MedChem Express (Monmouth Junction, NJ) or Chemietek (Indianapolis, IN) and dissolved in dimethyl sulfoxide (DMSO). In single-dose pharmacokinetics studies in human patients, maximum observed plasma concentrations for buparlisib and selumetinib were 2–5 μM (1–2 μg/mL) and 1.1–2.0 μM (0.5–0.9 μg/mL), respectively [39, 40]. Depending on experimental requirements, drugs were used at or above these clinically relevant dose ranges.
Immunoblots
Control (parental, GFP, and PIK3CAWT) and PIK3CAmut NHA and NHARAS cells were treated with either vehicle control (DMSO), buparlisib and/or selumetinib, or serum starved for 24 h. Proteins were isolated and immunoblots were performed as described [19, 24, 35]. Raw immunoblot images are shown in Supplemental Immunoblots. Each blot included both a molecular weight ladder and a reference standard composed of lysates of cultured TRP astrocytes. Quantification was performed using the following formula, where x is an individual blot and i is the target in question: [19, 24, 35]. N = 1–3 blots, mean is denoted in the corresponding figure legend. Bands annotated in red are omitted from final figures.
Cell growth
NHA and NHARAS were plated in triplicate or quadruplicate in 96-well tissue culture plates and absorbance (cell growth) was assessed using CellTiter 96 Aqueous One Cell Proliferation Assay (MTS, Promega, Madison, WI) according to manufacturer’s instructions. Relative absorbance was measured daily as described and fit to an exponential growth equation to calculate rate constants (k) and doubling times [ln(2)/k]. Differences in growth rate constants (k) were compared using the extra-sum-of-squares F test [35].
Cell migration
Migration rate across a cell-free gap was determined using culture inserts according to manufacturer’s instructions (Ibidi, Munich, Germany). Briefly, cells were imaged every 2 hours for 12 hours after creation of a cell free gap using a VistaVision inverted microscope equipped with a 4X objective and a DV-300 camera (VWR, Radnor, PA). Gap closure rates were calculated from 2–12 hours using linear regression and compared via ANCOVA.
Colony formation in soft agar
Colony formation was determined as described with minor modifications [25, 41, 42]. Briefly, cells were suspended in a mixture of DMEM/0.35% agarose (Denville Scientific INC., Holliston, MA) supplemented with 2.5% FBS and 14,000 cells were plated per well in 6-well plates. Cells were maintained for 4 weeks, fixed, and stained with 0.005% crystal violet in 70% ethanol. Plates were imaged on a Typhoon Trio (GE Healthcare) and colonies ≥ 30 μm2 were counted.
Mouse use
This study was carried out in strict accordance with the recommendation of the Guide for the Care and Use of Laboratory Animals of the National Institute of Health. The Institutional Animal Care and Use Committee of the University of North Carolina (Chapel Hill, NC) approved this study (Protocol #16–112). Animals were housed in a SPF facility in IVC cages with enrichment of nestlets and a shelter on corn cob bedding at a density of 2–5 animals per cage. Animals were kept on a 12-hour light/dark cycle at a temperature of 21o +/- 2o Celsius and were monitored daily by experienced laboratory staff following experimental initiation.
Orthotopic xenografts
Control and PIK3CAmut NHARAS lines were harvested by trypsinization, counted, and suspended in serum-free DMEM with 5% methyl cellulose. Male and female adult athymic (Foxn1nu/nu) nude mice (Charles River, Wilmington, MA; mean age ~3 months; N = 5–10 per group, mean = 9) were anesthetized with Avertin (250 mg/kg) and 2 x 105 GFP, PIK3CAWT, PIK3CAR88Q, PIK3CAE542K, or PIK3CAH1047R NHARAS cells were injected into the right basal ganglia of mice (N = 5–10 per group, mean = 9) using the coordinates 1, -2, and -4 mm (A, L, D) from bregma as previously described [19, 24]. Subjects received bupivacaine for local nerve block and a single dose of ketorolac for post-surgical analgesia. Animals were monitored daily for the onset of neurological symptoms (lethargy, hunching, seizures, paralysis, loss of righting reflex) and euthanized via CO2 asphyxiation within 24 hours of onset, immediately followed by brain tissue harvest. Symptoms were frequently severe at first observation due to rapid tumor progression. Animals did not die without euthanasia. Survival was determined by Kaplan-Meier analyses and was compared by log-rank tests.
Drug response
Dose response assays using MTS were performed and IC50 calculated as described [24, 35]. Synergy between MEKi and PI3Ki was determined by BLISS using Combenefit v1.31 [43].
Statistics
GraphPad Prism (La Jolla, CA) was used for statistical analyses. Error bars are SEM unless otherwise stated. P≤0.05 were considered significant.
Results
PIK3CAmut are frequent in GBM and are heterogeneously distributed across multiple encoded protein domains, including ABD, helical, and kinase (S1A Fig) [15, 16]. PTEN deletion or activating AKT mutations cooperate with activated MAPK signaling (hereafter MAPK) to promote tumorigenesis in preclinical glioma models [19, 22]. However, the role of PIK3CAmut in has not been examined in these models. To this end, we examined the 2 most frequent (hotspot) helical (E542K, E545K) and kinase (M1043V, H1047R) domain PIK3CAmut found in GBM and other cancer types (S1A–S1C Fig). ABD mutations are less prevalent in most cancers. Of these, R88Q is the most common and only recurrent mutation in GBM [4, 15]. We therefore evaluated it, as well as a second, C90Y. We transduced each of 6 PIK3CAmut into NHA and NHARAS via lentiviral vectors. Parental, GFP, or PIK3CAWT-transduced lines served as controls.
PIK3CAmut induce PI3K in vitro
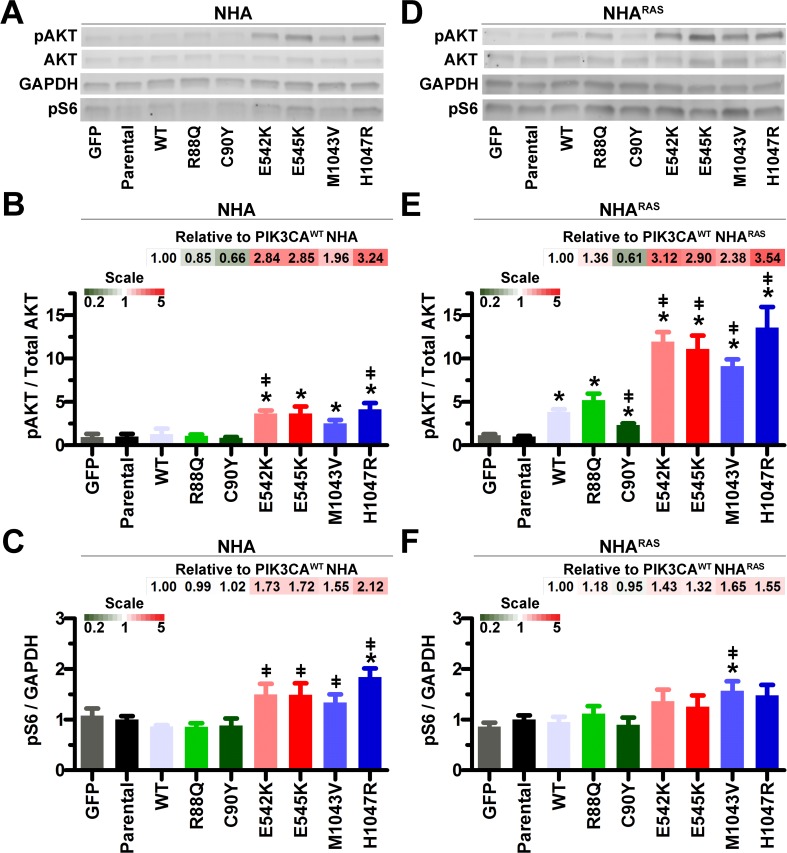
Expression of PIK3CAWT and all 6 PIK3CAmut was similar in both NHA and NHARAS (S2 Fig), suggesting that phenotypic differences would be attributable to PIK3CAmut examined. Neither PIK3CAWT nor ABD PIK3CAmut significantly activated proximal (pAKT) or distal (pS6) PI3K in serum-starved NHA (Fig 1A–1C). In contrast, E542K and H1047R significantly induced proximal and all 4 helical/kinase mutations induced distal PI3K.
Fig 1. Helical and kinase PIK3CAmut activate proximal PI3K.
Representative immunoblots (A) and quantification showed that proximal PI3K (pAKT) was increased by helical and kinase mutants in NHA (B). Distal PI3K (pS6) was only increased by H1047R (C) (*, P≤0.02). E542K and H1047R increased proximal PI3K and all helical and kinase mutants increased distal PI3Kcompared to PIK3CAWT NHA (ǂ, P≤0.04). Representative immunoblots (D) and quantification showed that proximal PI3K was increased by PIK3CAWT and all mutants (E). Helical and kinase mutants also increased proximal PI3K compared to PIK3CAWT NHARAS (ǂ, P≤0.007). Only M1043V increased distal PI3K compared to parental (*, P = 0.03) and PIK3CAWT (ǂ, P = 0.03) NHARAS (F). Bar graph data are set relative to parental lines (N = 4 biologic replicates). Fold changes in pAKT and pS6 relative to PIK3CAWT are shown as heatmaps.
We previously showed that mutant Kras cooperates with Pten deletion to activate PI3K in immortalized mouse astrocytes [19]. Therefore, we also determined the effects of PIK3CAmut on PI3K in NHARAS. PIK3CAWT and all PIK3CAmut increased proximal PI3K compared to parental NHARAS (Fig 1D and 1E). Furthermore, helical and kinase PIK3CAmut potentiated proximal PI3K more than PIK3CAWT. However, distal PI3K was only increased by M1043V in NHARAS (Fig 1D and 1F). Taken together, these results suggest that mutant RAS cooperated with ectopic expression of both PIK3CAWT and PIK3CAmut to increase proximal PI3K.
There is extensive cross-talk between PI3K and MAPK pathways [44]. We therefore determined the effects of PIK3CAmut on MAPK. Neither PIK3CAWT nor any of the PIK3CAmut significantly altered MAPK (pERK1/2) in NHA and NHARAS (S3 Fig). Thus, PIK3CAmut activated PI3K without affecting the MAPK pathway.
PIK3CAmut induce NHA proliferation in vitro
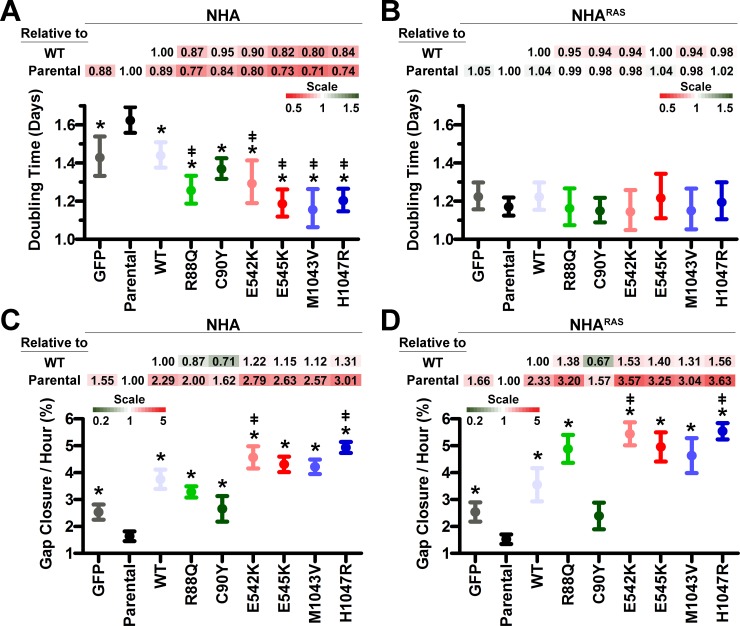
PIK3CAmut, particularly helical and kinase mutants, activated PI3K, suggesting that they may also promote cell growth (Fig 1). MTS assays using high-serum (10% FBS) showed that PIK3CAWT and a subset of PIK3CAmut slightly increased growth rate (≤15%) of rapidly-proliferating NHA (doubling times ≤ 1 day; S4 Fig). We therefore hypothesized that growth factor concentrations in high-serum media were masking the effects of PIK3CAmut on NHA growth. Indeed, MTS assays using low-serum (2.5% FBS) revealed increased proliferation of both GFP and PIK3CAWT NHA compared to parental cells (Fig 2A and S5A Fig). While all PIK3CAmut except C90Y increased proliferation compared to parental and PIK3CAWT NHA, proliferation rates were similar in all NHARAS lines (Fig 2B and S5B Fig). Taken together, these data suggest that PIK3CAmut promote astrocyte growth in the absence, but not presence, of mutant RAS.
Fig 2. PIK3CAmut potentiate proliferation and migration in vitro.
MTS assays showed that PIK3CAWT and all PIK3CAmut decreased doubling times of NHA (A), but not NHARAS (B) (*, P≤0.02 vs parental, S5A and S5B Fig). PIK3CAmut, except C90Y, decreased doubling times compared to PIK3CAWT NHA (ǂ, P≤0.03). Growth rates were analyzed by comparing k values. Error bars are 95% confidence intervals. PIK3CAWT and PIK3CAmut, except C90Y, increased migration of both NHA (C) and NHARAS (D) (*, P≤0.04, S5C and S5D Fig). E542K and H1047R also potentiated migration compared to PIK3CAWT NHA and NHARAS (ǂ, P≤0.005). Fold changes in doubling times and migration rates relative to parental and PIK3CAWT lines are shown as heatmaps.
PIK3CAmut induce migration in vitro
Complete surgical resection of GBM is precluded by its diffuse brain infiltration [45]. The PI3K pathway has an established role in migration [46]. We previously showed that Pten deletion increased migration of immortalized mouse astrocytes [19]. We therefore determined the effects of PIK3CAmut on migration of NHA and NHARAS using an in vitro gap closure assay. GFP, PIK3CAWT and all PIK3CAmut showed increased migration compared to parental NHA (Fig 2C and S5C Fig). PIK3CAWT and PIK3CAmut except C90Y migrated faster than GFP NHA (P≤0.04). Similarly, PIK3CAmut except C90Y migrated faster than GFP and parental NHARAS (P≤0.006, Fig 2D and S5D Fig). Moreover, E542K and H1047R PIK3CAmut migrated faster than NHA and NHARAS overexpressing PIK3CAWT (Fig 2C and 2D).
PIK3CAmut potentiate cellular transformation and tumorigenesis
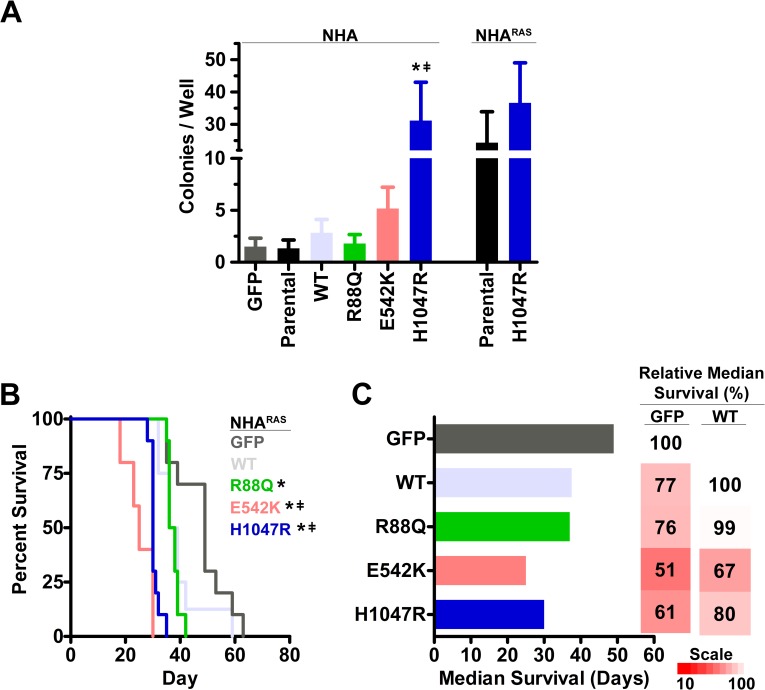
Anchorage-independent growth (colony formation in soft agar) is an established marker of cellular transformation [25, 41]. NHARAS, but not NHA, form colonies in vitro and develop high-grade tumors in orthotopic mouse xenograft models [25]. We therefore first determined whether PIK3CAmut promote NHA colony formation by selecting the most potent mutant in each domain (R88Q, E542K, H1047R) based on their effect on proximal PI3K, proliferation, and migration in NHA (Figs 1 and 2). Only H1047R induced colony formation relative to GFP and parental NHA (Fig 3A). Since this was the only mutation to potentiate NHA colony formation, we also tested its effect in NHARAS. However, no significant increase in colony formation was evident (Fig 3A).
Fig 3. Helical and kinase PIK3CAmut potentiate cellular transformation and tumorigenesis.
Only H1047R increased colony formation compared to parental (*, P = 0.03) and PIKCAWT (ǂ, P = 0.04) NHA (A). H1047R did not affect colony formation of NHARAS (P = 0.5). Orthotopic xenografts of GFP, PIK3CAWT, and PIK3CAmut NHARAS (BC). Median survival of mice with R88Q, E542K, or H1047R PIK3CAmut NHARAS was decreased compared to GFP control tumors (*, P≤0.003). E542K and H1047R PIK3CAmut also decreased survival compared to PIK3CAWT (ǂ, P≤0.002) and R88Q PIK3CAmut (P<0.0001). Fold changes in median survival relative to GFP and PIK3CAWT NHARAS are shown as heatmaps.
We next performed orthotopic xenografts of GFP, PIK3CAWT, and PIK3CAmut in NHARAS to determine whether PIK3CAmut potentiate tumorigenesis in vivo. Mice with R88Q, E542K, or H1047R PIK3CAmut NHARAS tumors died more quickly than mice with control GFP tumors (P≤0.003). Additionally, mice with E542 or H1047R PIK3CAmut tumors succumbed to disease more quickly than mice with tumors that overexpressed either PIK3CAWT (P≤0.002) or R88Q PIK3CAmut (P<0.0001; Fig 3B and 3C). Upon histopathological examination, tumor morphology was consistent across genotypes (S6 Fig). Taken together, these results indicate that both the mutated domain and concomitant mutant RAS influence the role of PIK3CAmut in gliomagenesis in vitro and in vivo.
PI3Ki efficacy is similar regardless of PIK3CAmut status
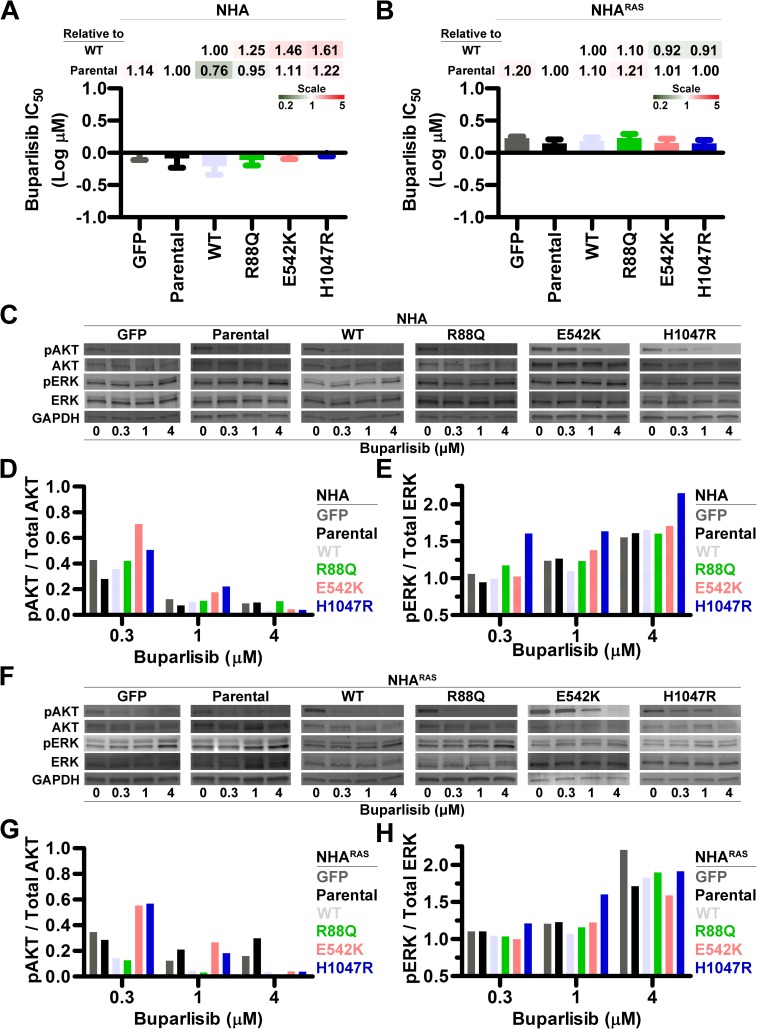
Neuro-oncology is transitioning towards precision medicine, wherein tumor mutation profiles are utilized to tailor targeted treatments [8, 47]. Knowing which oncogenic driver mutations influence targeted inhibitor response is a prerequisite. To this end, we determined the effects of PIK3CAmut on efficacy of the PI3Ki buparlisib in vitro. Buparlisib induced a dose-dependent decrease in growth of control and PIK3CAmut NHA and NHARAS (S7A and S7B Fig). High nanomolar IC50 were evident regardless of the specific PIK3CAmut (Fig 4A) but tended to be slightly higher in NHARAS (Fig 4B and S7C Fig). Buparlisib also induced G2/M cell cycle arrest in NHARAS lines regardless of PIK3CAmut status (S8 Fig).
Fig 4. PI3Ki inhibits growth and ablates PI3K regardless of PIK3CAmut status.
Buparlisib IC50 were similar regardless of PIK3CAmut status in NHA (A) and NHARAS (B) (S7 Fig). Fold changes in IC50 relative to parental and PIK3CAWT are shown as heatmaps. Representative immunoblots of control and PIK3CAmut NHA (C) and NHARAS (F) treated with buparlisib for 24 h. Immunoblot quantification (DEGH) demonstrated dose-dependent decreases in proximal PI3K (DG), with corresponding increases in MAPK (EH) in NHA (DE) and NHARAS (GH) lines (S10 Fig). Western blots were performed in either biological duplicates or triplicates.
PI3Ki ablate PI3K and potentiate MAPK
While PIK3CAmut did not alter PI3Ki efficacy in NHA and NHARAS in vitro (Fig 4A and 4B), they differentially activated PI3K (Fig 1). We therefore investigated whether PIK3CAmut influence PI3Ki-induced changes in pathway signaling. Buparlisib inhibited proximal (Fig 4C and 4D) and distal (S9 Fig) PI3K dose-dependently in control and PIK3CAmut NHA. We and others have shown that PI3Ki induce alternate MAPK activation in preclinical GBM models [35, 44, 48]. Indeed, buparlisib induced dose-dependent increases in MAPK in NHA, regardless of their PIK3CAmut status (Fig 4C and 4E).
Mutant RAS cooperated with PIK3CAWT and PIK3CAmut to potentiate activation of proximal PI3K (Fig 1), so we investigated whether RAS status influences PI3Ki-induced changes in PI3K and MAPK signaling. Buparlisib induced dose-dependent PI3K inhibition and MAPK activation in both control and PIK3CAmut NHARAS (Fig 4F–4H and S9C and S9D and S10 Figs). Proximal PI3K inhibition was least pronounced in helical and kinase PIK3CAmut lines at low buparlisib concentrations, demonstrating that higher PI3Ki doses are required to ablate PI3K in the presence of PIK3CAmut in NHARAS. These results also indicate that PIK3CAmut status does not influence alternate MAPK activation.
MEKi efficacy is independent of PIK3CAmut in NHA
Because PI3Ki promoted MAPK regardless of PIK3CA/RAS status, we determined efficacy of the MEKi selumetinib in control and PIK3CAmut NHA and NHARAS lines in vitro. Selumetinib caused gradual, dose-dependent decreases in growth (S11A and S11B Fig) and had similar IC50 in both parental NHA and NHARAS. While PIK3CAmut status influenced IC50 in neither NHA (Fig 5A) nor most NHARAS lines, C90Y and H1047R were slightly more resistant than parental NHARAS. (Fig 5B and S11C Fig). Thus, PIK3CAmut and mutant RAS had little to no effect on MEKi sensitivity.
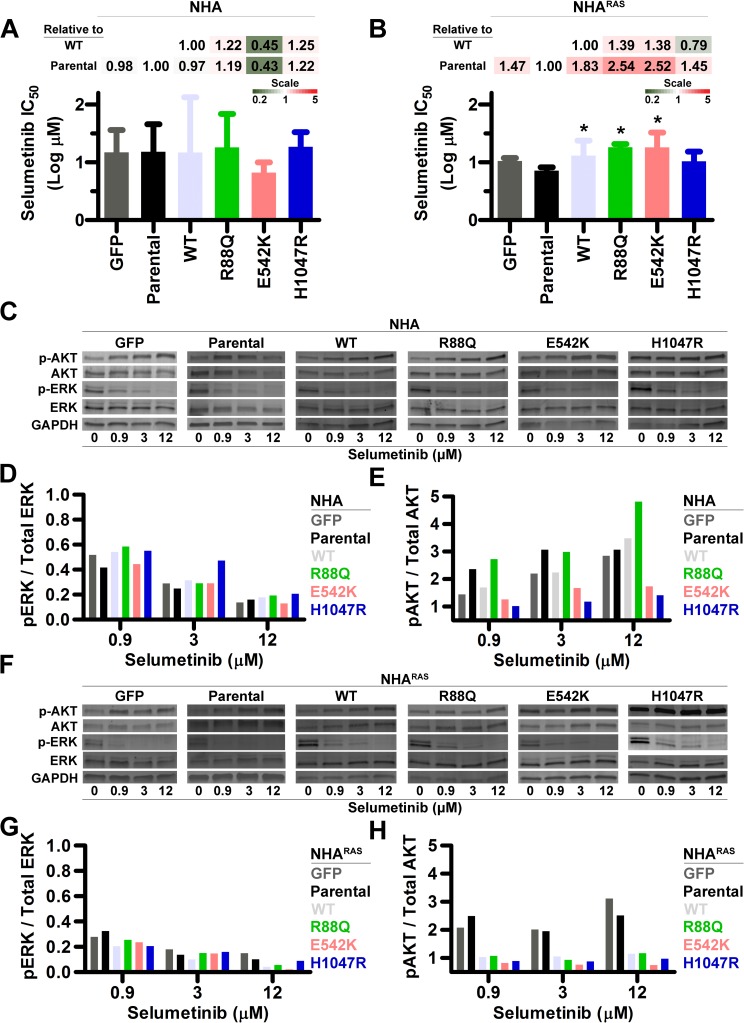
Fig 5. MEKi inhibits growth and ablates MAPK regardless of PIK3CAmut status.
Selumetinib IC50 were similar regardless of PIK3CAmut status in NHA (A), but slightly higher in most PIK3CAmut NHARAS compared to parental cells (*, P≤0.03) (B) (S11 Fig). Fold changes in IC50 relative to parental and PIK3CAWT lines are shown as heatmaps. Representative immunoblots of control and PIK3CAmut NHA (C) and NHARAS (F) treated with selumetinib for 24 h. Immunoblot quantification (DEGH) demonstrated dose-dependent decreases in MAPK in NHA (D) and NHARAS (G) lines. Although proximal PI3K was induced in control and PIK3CAmut NHA (E), it was only potentiated in GFP and parental NHARAS (H) (S12 Fig). Western blots were performed either 1 or 2 times per experiment (Mean = 1.8).
PIK3CAWT and PIK3CAmut influence MEKi-induced PI3K activation in NHARAS
Selumetinib inhibited MAPK in both control and PIK3CAmut NHA (Fig 5C) and induced dose-dependent decreases in pERK regardless of PIK3CAmut status (Fig 5C and 5D). We and others have shown that MEKi induces alternate PI3K activation in preclinical GBM models [35, 44, 49]. We extended these findings here, showing that selumetinib potentiated proximal PI3K 1.4-5-fold in control and PIK3CAmut NHA (Fig 5C and 5E). Induction in NHA was least pronounced with E542K and H1047R, the mutations that most potentiated tumorigenesis in NHARAS (Fig 3B and 3C).
Mutant RAS cooperated with PIK3CAWT and PIK3CAmut to promote activation of proximal PI3K (Fig 1). We therefore investigated whether PIK3CAmut influence MEKi-induced changes in MAPK and PI3K in NHARAS. PIK3CAmut status did not affect MAPK inhibition in selumetinib-treated NHARAS lines (Fig 5F and 5G and S12A and S12B Fig). Selumetinib induced alternate activation of proximal PI3K in GFP and parental NHARAS but ablated it in PI3KCAWT and all PIK3CAmut NHARAS (Fig 5F and 5H and S12A and S12C Fig). Taken together, these results indicate that ectopic PIK3CA expression in combination with mutant RAS prevents MEKi-induced alternate PI3K activation.
Dual PI3Ki/MEKi treatment is synergistic in PIK3CAmut NHA and NHARAS
We and others have shown that dual PI3Ki/MEKi efficacy is increased relative to treatment with either alone [35, 44, 48, 49]. However, the effects of GBM-associated mutations on PI3Ki/MEKi synergism remain unclear. To this end, we determined whether PIK3CAmut influence the effects of dual PI3Ki/MEKi treatment on NHA and NHARAS growth in vitro. Buparlisib plus selumetinib synergistically inhibited growth in control and PIK3CAmut NHA (Fig 6A). Synergy was highest in R88Q and E542K NHA relative to PIK3CAWT and H1047R.
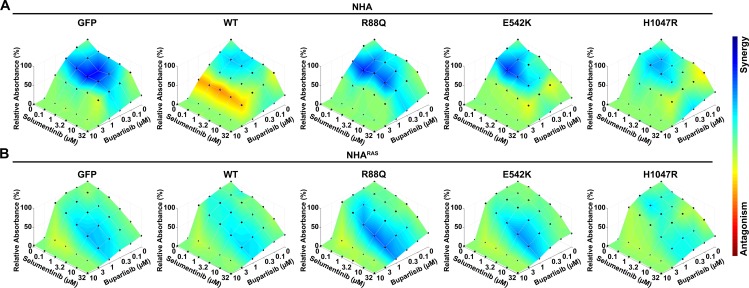
Fig 6. PI3Ki/MEKi synergism in vitro is influenced by PIK3CAmut and mutant RAS.
Buparlisib and selumetinib inhibited growth and were synergistic in control and PIK3CAmut NHA (A) and NHARAS (B). BLISS showed that synergy was most pronounced with high nanomolar buparlisib and low micromolar/high nanomolar selumetinib in NHA lines. In contrast, synergistic concentrations in NHARAS lines were generally most pronounced at both low micromolar buparlisib and selumetinib.
PIK3CAWT and PIK3CAmut marginally decreased MEKi efficacy in NHARAS (Fig 5B and S11B and S11C Fig). They also cooperated with mutant RAS to prevent MEKi-induced potentiation of proximal PI3K (Fig 5F–5H and S12 Fig). Therefore, both PIK3CAWT and PIK3CAmut may alter PI3Ki/MEKi synergism when combined with mutant RAS. Dual buparlisib/selumetinib treatment synergistically inhibited growth of all NHARAS lines (Fig 6B). However, synergism was most pronounced at higher drug concentrations in NHARAS relative to NHA lines. Furthermore, synergy was highest in R88Q, E542K, and M1043V mutant NHARAS (S13 Fig). Taken together, these data suggest that mutant RAS and PIK3CA alter PI3Ki/MEKi synergism.
Discussion
PIK3CAmut differentially activate PI3K and promote gliomagenesis
The vast majority of GBM harbor mutations in core PI3K pathway genes and/or upstream RTK [4]. Activation of PI3K via Pten deletion, PIK3R1 mutations, or constitutively active AKT mutants promotes tumorigenesis in glioma models [19–22]. Here, we determined the effects of ABD, helical, and kinase PIK3CAmut on gliomagenesis. Both helical and kinase PIK3CAmut potentiated PI3K, proliferation, and migration of NHA compared to parental and PIK3CAWT lines (Figs 1 and 2), but only H1047R kinase mutation potentiated NHA colony formation (Fig 3A).
We and others have shown that PI3K activation via Pten deletion or constitutively active AKT cooperates with MAPK activation to potentiate gliomagenesis [19, 22–24]. We extended these findings here by demonstrating that mutant RAS promoted PIK3CAWT- and PIK3CAmut-induced proximal PI3K (Fig 1). Unlike in NHA, PIK3CAmut did not increase proliferation of NHARAS, likely due to the rapid proliferation rate of parental cells (Fig 2A and 2B). H1047R PIK3CAmut also did not potentiate colony formation of NHARAS, likely because NHARAS cells are more aggressive and form colonies more readily and at higher density than NHA cells (Fig 3A). However, the E542K and H1047R PIK3CAmut potentiated malignancy of NHARAS in vivo compared to GFP and PIK3CAWT (Fig 3B and 3C). These results are consistent with previous findings that constitutively active AKT does not enhance proliferation or colony formation of NHARAS in vitro but promotes tumorigenesis in vivo [22]. These data suggest that E542K and H1047R PIK3CAmut promote in vivo gliomagenesis equally in the presence, but not absence, of mutant RAS.
In contrast to helical and kinase PIK3CA mutations, ABD PIK3CAmut did not increase PI3K, migration, or colony formation of NHA more than PIK3CAWT (Figs 1–3). Similar results were obtained with ABD PIK3CAmut in NHARAS. Moreover, R88Q PIK3CAmut did not promote tumorigenesis of NHARAS more than PIK3CAWT in vivo (Fig 3B and 3C). Taken together, these results demonstrate that the phenotypic consequences of ABD PIK3CAmut and ectopic over-expression of PIK3CAWT are similar. Furthermore, they suggest that ABD PIK3CAmut may be passenger mutations in GBM. However, ectopic expression of PIK3CAWT and PIK3CAmut may not fully recapitulate the effects of PIK3CAmut when expressed under its endogenous promoter. Furthermore, other cooperating mutations and/or cellular origin may influence the role of PIK3CAmut in gliomagenesis. Future work will be required to investigate the role of PIK3CAmut in other genetic and cellular contexts.
PIK3CAmut do not influence PI3Ki efficacy
The precision medicine initiative seeks to direct treatment with targeted inhibitors based on tumor mutation profiles [8, 47]. However, this requires an understanding of how oncogenic mutations influence drug response. Mutational activation of kinases can cause oncogene addiction, in which tumor cells become reliant upon the activated signaling pathway(s), and are thus highly sensitive to their inhibition [50]. Additionally, kinase mutations can alter drug affinity, thereby altering efficacy [51]. Buparlisib inhibits purified PIK3CAWT and the most common PIK3CAmut, E542K, E545K, and H1047R, with similar IC50 [52, 53]. Because these helical and kinase domain PIK3CAmut activated PI3K and promoted gliomagenesis, we hypothesized that they would also increase PI3Ki efficacy. However, higher buparlisib doses were required to ablate PI3K in cells expressing PIK3CAmut, particularly those in the helical and kinase domains, and these mutations did not influence PI3Ki efficacy in vitro (Fig 4). These results suggest that PIK3CAmut neither induce oncogene addiction nor enhance PI3Ki sensitivity. Whether they influence efficacy of isoform-specific PI3Ki or inhibitors of downstream kinases, such as AKT and mTOR, remains to be determined.
PIK3CAmut influence MEKi-induced PI3K activation and PI3Ki/MEKi synergism
We previously found that buparlisib induced widespread kinome changes, including MAPK activation, in immortalized murine astrocytes with Pten deletion and mutant Kras [35]. We expanded these findings here by demonstrating that buparlisib potentiated MAPK regardless of RAS/PIK3CA mutation status (Fig 4C–4H). PIK3CAmut also had minimal to no effect on sensitivity of NHA and NHARAS to MEKi in vitro (Fig 5A and 5B).
We and others have shown that MEKi promote PI3K in preclinical GBM models [35, 44, 49]. We found that selumetinib increased proximal PI3K in control and PIK3CAmut NHA, and in GFP and parental NHARAS (Fig 5C–5H). Interestingly, this increase was not apparent in PIK3CAWT and PIK3CAmut NHARAS (Fig 5F and 5H and S12A and S12C Fig). The mechanism by which ectopic PIK3CA expression in combination with mutant RAS alters MEKi response is unclear. A mutually inhibitory crosstalk between PI3K and MAPK is mediated by p70S6K in glioma stem cells [44]. MAPK inhibition induces PI3K in non-GBM cell lines via removal of a negative feedback loop on RTK [54]. Similarly, selumetinib induces widespread kinome changes in breast cancer models, including increased expression and activity of multiple RTK [55]. Taken together, these results suggest that PIK3CAWT and PIK3CAmut may cooperate with mutant RAS to alter MEKi-induced dynamic kinome changes, particularly as it pertains to PI3K activation.
Dual PI3Ki/MEKi treatment is effective in multiple preclinical GBM models [35, 44, 48, 49]. It remains unclear whether the underlying genetics of these models influence drug synergism. We therefore determined if PIK3CAmut affected PI3Ki/MEKi synergism in the presence and absence of mutant RAS. Consistent with other GBM models, we found that dual buparlisib/selumetinib treatment was synergistic in NHA and NHARAS lines (Fig 6). However, RAS/PIK3CAmut status influenced drug response. Higher concentrations of buparlisib and selumetinib were required to maximize synergism in NHARAS lines compared to NHA. Furthermore, synergy was generally greater in R88Q and E542K NHA and NHARAS compared to those with either PIK3CAWT or H1047R. Taken together, these results suggest that GBM patients with helical PIK3CAmut may be most sensitive to dual PI3Ki/MEKi treatment. Given that ABD PIK3CAmut showed no significant tumorigenic effects in vitro and in vivo, their utility in predicting PI3Ki/MEKi synergy remains questionable.
Conclusion
Defining the role of frequently occurring mutations in GBM pathogenesis and drug response can aid identification of predictive biomarkers. Our results demonstrate that PIK3CAmut differentially promote gliomagenesis and do not predict PI3Ki sensitivity but do impact PI3Ki/MEKi synergism.
Supporting information
Lollipop plot of PIK3CA missense (green), in-frame deletion (brown), and truncating mutations (black) in GBM (A) and all published TCGA datasets (B). PIK3CA missense mutations investigated here are indicated. PIK3CA point mutations were evident in 10.3% of GBM cases from the TCGA (N = 273), with each mutation investigated here representing ≤1% of total mutations. Data were downloaded from cBioPortal (http://www.cbioportal.org/) on March 10, 2017. Ribbon diagram of PIK3CA with mutations investigated highlighted (C) (R88Q = light green; C90Y = dark green; E542K = pink; E545K = red; M1043V = purple; H1047R = blue). Model was generated in PyMOL. (Schrödinger, New York City, NY) using a script downloaded from cBioPortal.13,14
(TIF)
Representative immunoblots (AC) and quantification (BD) of HA-tagged PIK3CA showed that PIK3CAWT and PIK3CAmut were expressed at similar levels in NHA (BC) and NHARAS (CD) (ANOVA, P≥0.3). Bar graph data were set relative to PIK3CAWT lines (N = 3–4 biologic replicates, Mean = 3.5).
(TIF)
Representative immunoblots (AC) and quantification (BD) showed that PIK3CAmut did not alter MAPK (phosphorylation of ERK1/2, pERK) in either NHA (AB) or NHARAS (CD) (P≥0.93). Bar graph data were set relative to parental lines (N = 3–4 biologic replicates, Mean = 3.5). Fold changes in pERK relative to PIK3CAWT lines are shown as heatmaps.
(TIF)
MTS assays (A) showed that PIK3CAWT and a subset of PIK3CAmut slightly increased growth (reduced doubling times) compared to parental NHA (*, P≤0.03) when grown in media containing high (10%) FBS. (B). R88Q growth was slightly slower than PIK3CAWT NHA (ǂ, P = 0.01). Statistical analyses of growth rates were performed by comparing k values. Fold changes in doubling times relative to parental and PIK3CAWT lines are shown as heatmaps. Error bars in B are 95% confidence intervals.
(TIF)
Growth of control and PIK3CAmut NHA (A) and NHARAS (B) (Fig 2A and 2B). Growth was determined by assessing changes in relative absorbance daily by MTS. Migration of control and PIK3CA mutant NHA (C) and NHARAS (D) across a gap (Fig 2C and 2D).
(TIF)
Hematoxylin and eosin staining of tumors from PIK3CAWT (A), R88Q (B), E542K (C), and H1047R (D) PIK3CAmut mice revealed malignant histopathologic features typical of human gliomas, including cytologic and nuclear atypia, tumor giant cells, and mitotic figures (white arrows). Scale bar = 100 μm.
(TIF)
MTS assays showed that buparlisib caused dose-dependent decreases in growth of control and PIK3CAmut NHA (A) and NHARAS (B). Buparlisib IC50 were similar between control and all 6 PIK3CAmut NHARAS (C).
(TIF)
Micromolar doses of buparlisib induced G2/M cell cycle arrest within 48 h in control and PIK3CAmut NHARAS.
(TIF)
Representative immunoblots of control and PIK3CAmut NHA (A) and NHARAS (C) 24 h after buparlisib treatment. Immunoblot quantification demonstrated dose-dependent inhibition of distal PI3K in all NHA (B) and NHARAS (D) lines (N = 1–3 biologic replicates, Mean = 1.7).
(TIF)
Representative immunoblots (A) and quantification of proximal PI3K (B) and MAPK (C) showed that within 24 h, buparlisib induced dose-dependent inhibition of PI3K signaling, with concurrent induction of MAPK in parental, GFP, PIK3CAWT, and all 6 PIK3CAmut NHARAS lines (N = 2–3 biologic replicates, Mean = 2.7).
(TIF)
MTS assays showed that selumetinib caused dose-dependent decreases in growth of control and PIK3CAmut NHA (A) and NHARAS (B). Selumetinib IC50 were slightly increased by PIK3CAWT and all PIK3CAmut, except C90Y and H1047R, compared to parental NHARAS (*, P≤0.03) (C).
(TIF)
Representative immunoblots (A) and quantification of MAPK (B) and proximal PI3K (C) showed that selumetinib caused dose-dependent inhibition of MAPK regardless of PIKCAWT or PIK3CAmut status, but concurrent induction of proximal PI3K only occurred in parental and GFP NHARAS (N = 2–3 biologic replicates, Mean = 2.7).
(TIF)
Buparlisib and selumetinib were synergistic in parental and PIK3CAmut NHARAS (Fig 6B)
(TIF)
(DOCX)
(PPTX)
Acknowledgments
We thank Russell O. Pieper for providing NHA and NHARAS cell lines and UNC TPL for histology assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
CRM was supported by the UNC University Cancer Research Fund (UCRF), National Center for Advancing Translational Sciences (UL1TR001111, subaward 550KR61332), and National Cancer Institute (R01CA204136). RSM is a Robert H. Wagner Scholar and AKS is a Bill Sykes Scholar in Pathobiology and Translational Science. RSM was supported by the UNC Graduate Training Program in Translational Medicine. Translational Pathology Laboratory (TPL) is supported by National Cancer Institute (P30CA016086), National Institute of Environmental Health Sciences (P30ES010126), and UCRF.
References
- 1.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, et al. (2017) CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol 19: v1–v88. doi: 10.1093/neuonc/nox158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131: 803–820. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987–996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. (2013) The somatic genomic landscape of glioblastoma. Cell 155: 462–477. doi: 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. doi: 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110. doi: 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloughesy TF, Cavenee WK, Mischel PS (2014) Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol 9: 1–25. doi: 10.1146/annurev-pathol-011110-130324 [DOI] [PubMed] [Google Scholar]
- 8.Mendelsohn J (2013) Personalizing oncology: perspectives and prospects. J Clin Oncol 31: 1904–1911. doi: 10.1200/JCO.2012.45.3605 [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 10.Courtney KD, Corcoran RB, Engelman JA (2010) The PI3K pathway as drug target in human cancer. J Clin Oncol 28: 1075–1083. doi: 10.1200/JCO.2009.25.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fruman DA, Rommel C (2014) PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 13: 140–156. doi: 10.1038/nrd4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT (2017) The PI3K pathway in human disease. Cell 170: 605–635. doi: 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorpe LM, Yuzugullu H, Zhao JJ (2015) PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 15: 7–24. doi: 10.1038/nrc3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klempner SJ, Myers AP, Cantley LC (2013) What a tangled web we weave: emerging resistance mechanisms to inhibition of the phosphoinositide 3-kinase pathway. Cancer Discov 3: 1345–1354. doi: 10.1158/2159-8290.CD-13-0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1 doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen PY, Lee EQ, Reardon DA, Ligon KL, Alfred Yung WK (2012) Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro Oncol 14: 819–829. doi: 10.1093/neuonc/nos117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, et al. (2011) Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19: 305–316. doi: 10.1016/j.ccr.2011.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitucci M, Karpinich NO, Bash RE, Werneke AM, Schmid RS, White KK, et al. (2013) Cooperativity between MAPK and PI3K signaling activation is required for glioblastoma pathogenesis. Neuro Oncol 15: 1317–1329. doi: 10.1093/neuonc/not084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Y, Zhang Q, Kutlu B, Difilippantonio S, Bash R, Gilbert D, et al. (2013) Evolutionary etiology of high-grade astrocytomas. Proc Natl Acad Sci U S A 110: 17933–17938. doi: 10.1073/pnas.1317026110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quayle SN, Lee JY, Cheung LW, Ding L, Wiedemeyer R, Dewan RW, et al. (2012) Somatic mutations of PIK3R1 promote gliomagenesis. PLoS One 7: e49466 doi: 10.1371/journal.pone.0049466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonoda Y, Ozawa T, Aldape KD, Deen DF, Berger MS, Pieper RO (2001) Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res 61: 6674–6678. [PubMed] [Google Scholar]
- 23.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN (2000) Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 25: 55–57. doi: 10.1038/75596 [DOI] [PubMed] [Google Scholar]
- 24.Schmid RS, Simon JM, Vitucci M, McNeill RS, Bash RE, Werneke AM, et al. (2016) Core pathway mutations induce de-differentiation of murine astrocytes into glioblastoma stem cells that are sensitive to radiation but resistant to temozolomide. Neuro Oncol 18: 962–973. doi: 10.1093/neuonc/nov321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, et al. (2001) Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res 61: 4956–4960. [PubMed] [Google Scholar]
- 26.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, et al. (2007) The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science 318: 1744–1748. doi: 10.1126/science.1150799 [DOI] [PubMed] [Google Scholar]
- 27.Bader AG, Kang S, Vogt PK (2006) Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A 103: 1475–1479. doi: 10.1073/pnas.0510857103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sanchez V, et al. (2013) Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci U S A 110: 14372–14377. doi: 10.1073/pnas.1303204110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koren S, Bentires-Alj M (2013) Mouse models of PIK3CA mutations: one mutation initiates heterogeneous mammary tumors. FEBS J 280: 2758–2765. doi: 10.1111/febs.12175 [DOI] [PubMed] [Google Scholar]
- 30.Filbin MG, Dabral SK, Pazyra-Murphy MF, Ramkissoon S, Kung AL, Pak E, et al. (2013) Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med 19: 1518–1523. doi: 10.1038/nm.3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wachsberger PR, Lawrence YR, Liu Y, Rice B, Feo N, Leiby B, et al. (2014) Hsp90 inhibition enhances PI-3 kinase inhibition and radiosensitivity in glioblastoma. J Cancer Res Clin Oncol 140: 573–582. doi: 10.1007/s00432-014-1594-6 [DOI] [PubMed] [Google Scholar]
- 32.Speranza MC, Nowicki MO, Behera P, Cho CF, Chiocca EA, Lawler SE (2016) BKM-120 (Buparlisib): A Phosphatidyl-Inositol-3 Kinase Inhibitor with Anti-Invasive Properties in Glioblastoma. Sci Rep 6: 20189 doi: 10.1038/srep20189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Netland IA, Forde HE, Sleire L, Leiss L, Rahman MA, Skeie BS, et al. (2016) Treatment with the PI3K inhibitor buparlisib (NVP-BKM120) suppresses the growth of established patient-derived GBM xenografts and prolongs survival in nude rats. J Neurooncol 129: 57–66. doi: 10.1007/s11060-016-2158-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Y, Dai Y, Grant S, Dent P (2012) Enhancing CHK1 inhibitor lethality in glioblastoma. Cancer Biol Ther 13: 379–388. doi: 10.4161/cbt.19240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeill RS, Canoutas DA, Stuhlmiller TJ, Dhruv HD, Irvin DM, Bash RE, et al. (2017) Combination therapy with potent PI3K and MAPK inhibitors overcomes adaptive kinome resistance to single agents in preclinical models of glioblastoma. Neuro Oncol 19: 1469–1480. doi: 10.1093/neuonc/nox044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, et al. (2009) A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4: e6529 doi: 10.1371/journal.pone.0006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM (2005) The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A 102: 18443–18448. doi: 10.1073/pnas.0508988102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sowa ME, Bennett EJ, Gygi SP, Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403. doi: 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodon J, Brana I, Siu LL, De Jonge MJ, Homji N, Mills D, et al. (2014) Phase I dose-escalation and -expansion study of buparlisib (BKM120), an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. Invest New Drugs 32: 670–681. doi: 10.1007/s10637-014-0082-9 [DOI] [PubMed] [Google Scholar]
- 40.Dymond AW, Martin P, So K, Huang Y, Severin P, Holmes V, et al. (2017) Pharmacokinetics of a single oral dose of the MEK1/2 inhibitor selumetinib in subjects with end-stage renal disease or varying degrees of hepatic impairment compared with healthy subjects. J Clin Pharmacol 57: 592–605. doi: 10.1002/jcph.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. (2014) The soft agar colony formation assay. J Vis Exp: e51998 doi: 10.3791/51998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNeill RS, Schmid RS, Bash RE, Vitucci M, White KK, Werneke AM, et al. (2014) Modeling astrocytoma pathogenesis in vitro and in vivo using cortical astrocytes or neural stem cells from conditional, genetically engineered mice. J Vis Exp: e51763 doi: 10.3791/51763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. (2016) Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 32: 2866–2868. doi: 10.1093/bioinformatics/btw230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunayama J, Matsuda K, Sato A, Tachibana K, Suzuki K, Narita Y, et al. (2010) Crosstalk between the PI3K/mTOR and MEK/ERK pathways involved in the maintenance of self-renewal and tumorigenicity of glioblastoma stem-like cells. Stem Cells 28: 1930–1939. doi: 10.1002/stem.521 [DOI] [PubMed] [Google Scholar]
- 45.Giese A, Bjerkvig R, Berens ME, Westphal M (2003) Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol 21: 1624–1636. doi: 10.1200/JCO.2003.05.063 [DOI] [PubMed] [Google Scholar]
- 46.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 11: 329–341. doi: 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- 47.Prados MD, Byron SA, Tran NL, Phillips JJ, Molinaro AM, Ligon KL, et al. (2015) Toward precision medicine in glioblastoma: the promise and the challenges. Neuro Oncol 17: 1051–1063. doi: 10.1093/neuonc/nov031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Meskini R, Iacovelli AJ, Kulaga A, Gumprecht M, Martin PL, Baran M, et al. (2015) A preclinical orthotopic model for glioblastoma recapitulates key features of human tumors and demonstrates sensitivity to a combination of MEK and PI3K pathway inhibitors. Dis Model Mech 8: 45–56. doi: 10.1242/dmm.018168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.See WL, Tan IL, Mukherjee J, Nicolaides T, Pieper RO (2012) Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res 72: 3350–3359. doi: 10.1158/0008-5472.CAN-12-0334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torti D, Trusolino L (2011) Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: promises and perils. EMBO Mol Med 3: 623–636. doi: 10.1002/emmm.201100176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barouch-Bentov R, Sauer K (2011) Mechanisms of drug resistance in kinases. Expert Opin Investig Drugs 20: 153–208. doi: 10.1517/13543784.2011.546344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koul D, Fu J, Shen R, LaFortune TA, Wang S, Tiao N, et al. (2012) Antitumor activity of NVP-BKM120—a selective pan class I PI3 kinase inhibitor showed differential forms of cell death based on p53 status of glioma cells. Clin Cancer Res 18: 184–195. doi: 10.1158/1078-0432.CCR-11-1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maira SM, Pecchi S, Huang A, Burger M, Knapp M, Sterker D, et al. (2012) Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther 11: 317–328. doi: 10.1158/1535-7163.MCT-11-0474 [DOI] [PubMed] [Google Scholar]
- 54.Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, et al. (2012) MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res 72: 3228–3237. doi: 10.1158/0008-5472.CAN-11-3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. (2012) Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149: 307–321. doi: 10.1016/j.cell.2012.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lollipop plot of PIK3CA missense (green), in-frame deletion (brown), and truncating mutations (black) in GBM (A) and all published TCGA datasets (B). PIK3CA missense mutations investigated here are indicated. PIK3CA point mutations were evident in 10.3% of GBM cases from the TCGA (N = 273), with each mutation investigated here representing ≤1% of total mutations. Data were downloaded from cBioPortal (http://www.cbioportal.org/) on March 10, 2017. Ribbon diagram of PIK3CA with mutations investigated highlighted (C) (R88Q = light green; C90Y = dark green; E542K = pink; E545K = red; M1043V = purple; H1047R = blue). Model was generated in PyMOL. (Schrödinger, New York City, NY) using a script downloaded from cBioPortal.13,14
(TIF)
Representative immunoblots (AC) and quantification (BD) of HA-tagged PIK3CA showed that PIK3CAWT and PIK3CAmut were expressed at similar levels in NHA (BC) and NHARAS (CD) (ANOVA, P≥0.3). Bar graph data were set relative to PIK3CAWT lines (N = 3–4 biologic replicates, Mean = 3.5).
(TIF)
Representative immunoblots (AC) and quantification (BD) showed that PIK3CAmut did not alter MAPK (phosphorylation of ERK1/2, pERK) in either NHA (AB) or NHARAS (CD) (P≥0.93). Bar graph data were set relative to parental lines (N = 3–4 biologic replicates, Mean = 3.5). Fold changes in pERK relative to PIK3CAWT lines are shown as heatmaps.
(TIF)
MTS assays (A) showed that PIK3CAWT and a subset of PIK3CAmut slightly increased growth (reduced doubling times) compared to parental NHA (*, P≤0.03) when grown in media containing high (10%) FBS. (B). R88Q growth was slightly slower than PIK3CAWT NHA (ǂ, P = 0.01). Statistical analyses of growth rates were performed by comparing k values. Fold changes in doubling times relative to parental and PIK3CAWT lines are shown as heatmaps. Error bars in B are 95% confidence intervals.
(TIF)
Growth of control and PIK3CAmut NHA (A) and NHARAS (B) (Fig 2A and 2B). Growth was determined by assessing changes in relative absorbance daily by MTS. Migration of control and PIK3CA mutant NHA (C) and NHARAS (D) across a gap (Fig 2C and 2D).
(TIF)
Hematoxylin and eosin staining of tumors from PIK3CAWT (A), R88Q (B), E542K (C), and H1047R (D) PIK3CAmut mice revealed malignant histopathologic features typical of human gliomas, including cytologic and nuclear atypia, tumor giant cells, and mitotic figures (white arrows). Scale bar = 100 μm.
(TIF)
MTS assays showed that buparlisib caused dose-dependent decreases in growth of control and PIK3CAmut NHA (A) and NHARAS (B). Buparlisib IC50 were similar between control and all 6 PIK3CAmut NHARAS (C).
(TIF)
Micromolar doses of buparlisib induced G2/M cell cycle arrest within 48 h in control and PIK3CAmut NHARAS.
(TIF)
Representative immunoblots of control and PIK3CAmut NHA (A) and NHARAS (C) 24 h after buparlisib treatment. Immunoblot quantification demonstrated dose-dependent inhibition of distal PI3K in all NHA (B) and NHARAS (D) lines (N = 1–3 biologic replicates, Mean = 1.7).
(TIF)
Representative immunoblots (A) and quantification of proximal PI3K (B) and MAPK (C) showed that within 24 h, buparlisib induced dose-dependent inhibition of PI3K signaling, with concurrent induction of MAPK in parental, GFP, PIK3CAWT, and all 6 PIK3CAmut NHARAS lines (N = 2–3 biologic replicates, Mean = 2.7).
(TIF)
MTS assays showed that selumetinib caused dose-dependent decreases in growth of control and PIK3CAmut NHA (A) and NHARAS (B). Selumetinib IC50 were slightly increased by PIK3CAWT and all PIK3CAmut, except C90Y and H1047R, compared to parental NHARAS (*, P≤0.03) (C).
(TIF)
Representative immunoblots (A) and quantification of MAPK (B) and proximal PI3K (C) showed that selumetinib caused dose-dependent inhibition of MAPK regardless of PIKCAWT or PIK3CAmut status, but concurrent induction of proximal PI3K only occurred in parental and GFP NHARAS (N = 2–3 biologic replicates, Mean = 2.7).
(TIF)
Buparlisib and selumetinib were synergistic in parental and PIK3CAmut NHARAS (Fig 6B)
(TIF)
(DOCX)
(PPTX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.