Figure 6. Accelerated signs of in vitro T cell exhaustion in absence of ZFP36.
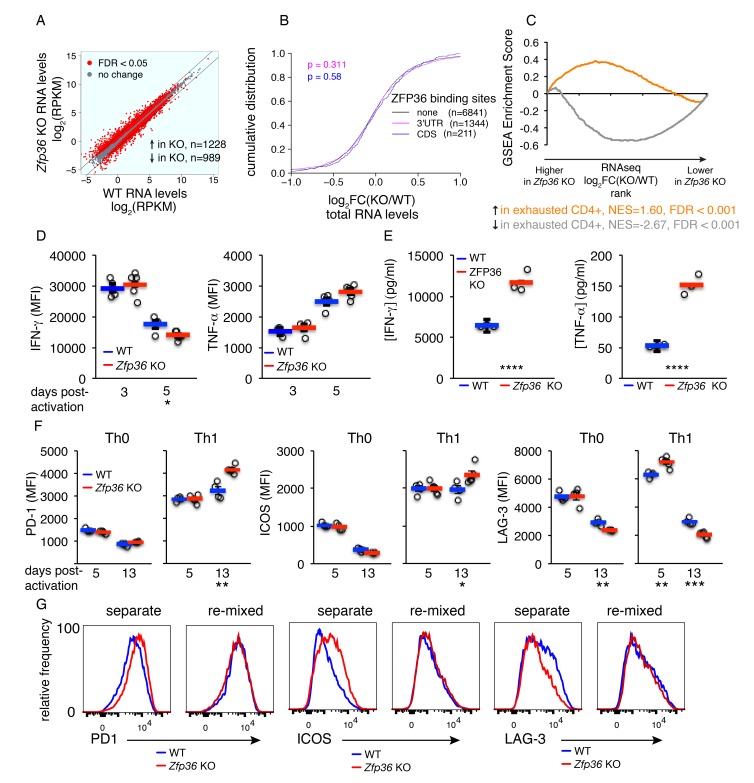
(A) Log2-transformed RPKM values from Zfp36 KO versus WT CD4 +Th1 cell RNAseq 72 hr post-activation, with red indicating differential expression (FDR < 0.05). Lines mark 2-fold changes. RNAseq data represent one experiment with three biological replicates per condition. (B) Log2-transformed fold-changes (KO/WT) plotted as a CDF, for mRNAs with 3’UTR, CDS, or no significant ZFP36 HITS-CLIP. Numbers of mRNAs in each category (n) and p-values from KS tests are indicated. (C) The gene expression profile in Zfp36 KO CD4 +T cells 72 hr post-activation was compared to reported profiles of CD4 +T cell exhaustion using GSEA. Upregulated (orange) and downregulated (gray) gene sets in exhausted T cells showed strong overlap with corresponding sets from Zfp36 KO T cells (FDR < 0.001, hypergeometric test). (D) IFN-γ and TNF-α measured by ICS 3 and 5 days after activation of naïve CD4 +T cells. (E) IFN-γ and TNF-α in culture supernatants 3 and 5 days after activation of naïve CD4 +T cells. (F) PD-1, ICOS, and LAG-3 expression 5 and 13 days after activation under Th0 or Th1 conditions. (D–F) show mean ± S.E.M.; circles are individual mice (n = 3–5 per genotype). (G) Measurements as in (F) for Zfp36 KO and WT CD4 +T cells derived from mixed BM chimeras. Cells were activated under Th1 conditions for 13 days, either separately or mixed 1:1. For (D–G), one representative experiment of two performed is shown. Results of two-tailed t-tests: *=p < 0.05; **=p < 0.01; ***=p < 0.001; ****=p < 0.0001.