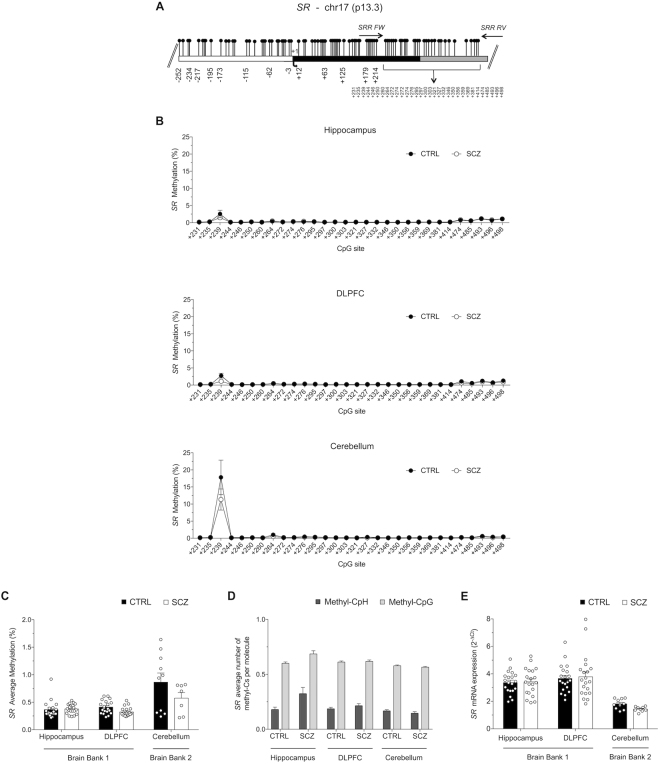
Figure 4.
SR methylation and expression analyses. (A) CpG-rich SR gene structure is reported with the white box, black box and grey box representing the putative regulatory upstream region, first exon and first intron, respectively. Transcriptional start site is indicated by the black arrow. Gene sequence was retrieved by Ensemble database accession number: SRR ENSG00000167720. (B) Average methylation at single CpG sites for each brain region is denoted in both groups (CTRL and SCZ). (C) SR methylation levels in HIPP, DLPFC and CB areas are shown for controls and schizophrenia patients. (D) The level of methylation at non-CpG and CpG sites in each molecule is shown. The percent values of non-CpG methylation is reported in Table S2. (E) SR mRNA expression levels in the hippocampus, DLPFC and cerebellum of control and schizophrenia patients were detected by quantitative RT-PCR and expressed as 2−ΔCt. All data are shown as the average ± standard error of the mean (SEM).