Fig E2.
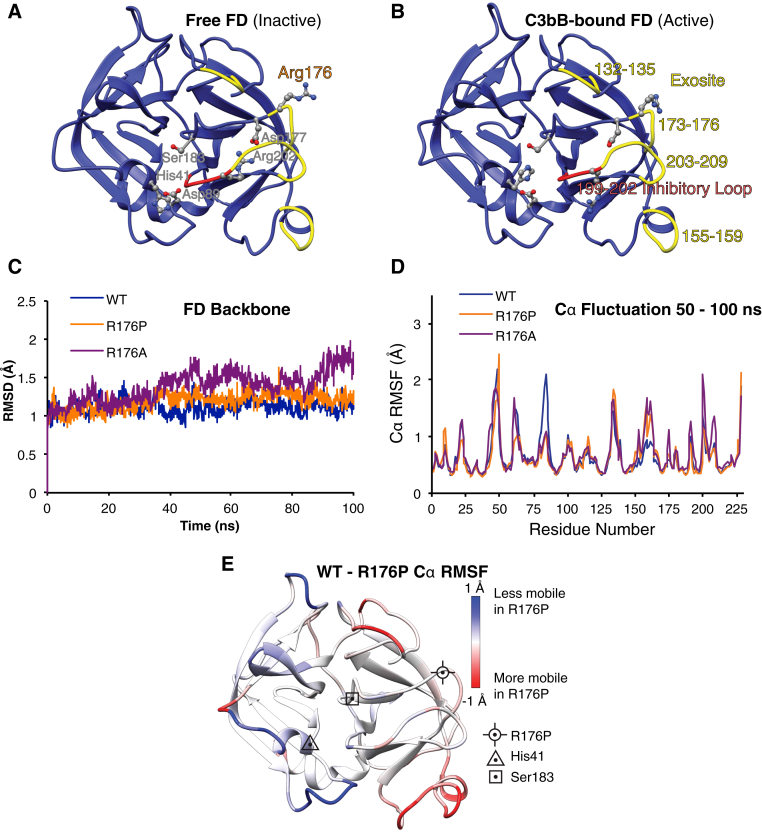
Mutation R176P stabilizes the self-inhibited state of FD. A, Structure of free FDE3 (PDB ID: 2XW9) showing the catalytic triad (Ser183-His41-Asp89) in an inactive conformation stabilized by the self-inhibitory loop 199-202 (red) and an ion bridge between Asp177 and Agr202. The exosite loops are shown in yellow. B, Structure of C3bB-bound FDE3 (PDB ID: 2XWB) omitting the C3b and FB components. FD exosite loops retain a conformation similar to that of unbound FD. C, WT, R176P, and R176A structures were stable over 100 nanoseconds of unrestrained molecular dynamics simulation with explicit solvent. D, Root mean square fluctuation (RMSF) in WT and mutant FD over the second half of the trajectory. E, Differences in WT versus R176 RMSF mapped to the FD structured. MD predicted increased mobility in exosite loops, notably 155-167, and decreased mobility in loops carrying the catalytic His41 and Asp89 residues.