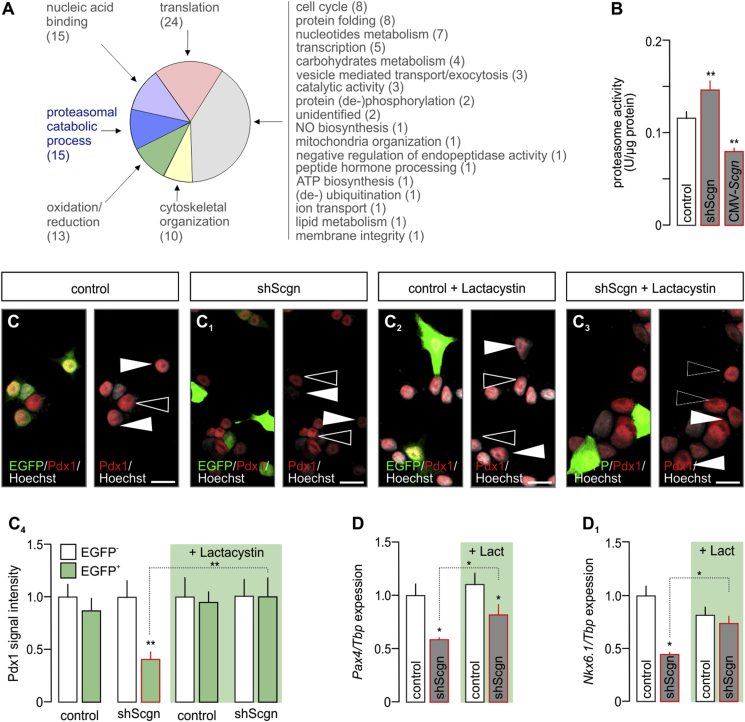
Figure 5.
Secretagogin controls Pdx1 by inhibiting the 26S proteasome. (A) Secretagogin's molecular interactome at E13.5. Recombinant His6-tagged secretagogin was used as bait with data adhering to gene ontology (GO) classifiers. Bracketed numbers refer to the number of proteins per GO cluster. The identity, discovery rate and peptide fragmentation data for all interacting proteins from our LC-MS/MS analyses are listed in Table S2. (B) Scgn silencing by shRNA (48 h) increases, whereas Scgn overexpression brought about by CMV-Scgn (48 h) reduces proteasome activity. (C-C4) Proteasome inhibition by lactacystin (5 μM, 6 h) rescues Pdx1 in shScgn-transfected INS-1E cells (EGFP+). Representative images are shown. Hoechst 33,342 was used as nuclear counterstain (pseudo-colored in gray). Scale bars = 5 μm. (C4) Quantitative analysis of Pdx1 signal intensity in lactacystin-treated INS-1E cells transfected with either mock or shScgn constructs. EGFP (arrowheads) indicates successful transfection. Open arrowheads point to non-transfected cells. (D–D2) Proteasome inhibition by lactacystin (‘Lact.’, 5 μM, 6 h) rescues Pax4 (D) and Nkx6.1 (D1) expression levels otherwise down-regulated by shScgn. Tbp mRNA level was used to normalize gene expression. Quantitative data from triplicate experiments were normalized to those in control and expressed as means ± s.d.; n ≥ 30 cells/group; *p < 0.05 (pair-wise comparisons after two-way ANOVA).