Abstract
Background
Increased coffee intake is associated with reduced serum urate concentrations and lower risk of gout. Specific alleles of the GCKR, ABCG2, MLXIPL, and CYP1A2 genes have been associated with both reduced coffee intake and increased serum urate in separate genome-wide association studies (GWAS). The aim of this study was to determine whether these single nucleotide polymorphisms (SNPs) influence the risk of gout through their effects on coffee consumption.
Methods
This research was conducted using the UK Biobank Resource. Data were available for 130,966 European participants aged 40–69 years. Gout status and coffee intake were tested for association with four urate-associated SNPs: GCKR (rs1260326), ABCG2 (rs2231142), MLXIPL (rs1178977), and CYP1A2 (rs2472297). Multiple regression and path analysis were used to examine whether coffee consumption mediated the effect of the SNPs on gout risk.
Results
Coffee consumption was inversely associated with gout (multivariate adjusted odds ratio (95% confidence interval (CI)) for any coffee consumption 0.75 (0.67–0.84, P = 9 × 10−7)). There was also evidence of a dose-effect with multivariate adjusted odds ratio (95% CI) per cup consumed per day of 0.85 (0.82–0.87, P = 9 × 10−32). The urate-increasing GCKR, ABCG2, MLXIPL, and CYP1A2 alleles were associated with reduced daily coffee consumption, with the strongest associations for CYP1A2 (beta −0.30, P = 8 × 10−40), and MLXIPL (beta −0.17, P = 3 × 10−8), and weaker associations for GCKR (beta −0.07, P = 3 × 10−10) and ABCG2 (beta −0.09, P = 2 × 10−9). The urate-increasing GCKR and ABCG2 alleles were associated with gout (multivariate adjusted p < 5 × 10−8 for both), but the urate-increasing MLXIPL and CYP1A2 alleles were not. In mediation analysis, the direct effects of GCKR and ABCG2 accounted for most of the total effect on gout risk, with much smaller indirect effects mediated by coffee consumption.
Conclusion
Coffee consumption is inversely associated with risk of gout. Although alleles at several SNPs associate with both lower coffee consumption and higher risk of gout, these SNPs largely influence gout risk directly, rather than indirectly through effects on coffee consumption.
Electronic supplementary material
The online version of this article (10.1186/s13075-018-1629-5) contains supplementary material, which is available to authorized users.
Keywords: Gout, Coffee, Genetics, Diet, Urate
Background
Gout is a common inflammatory arthritis characterised by deposition of monosodium urate (MSU) crystals in joints and other tissues [1]. Hyperuricaemia is a key checkpoint in MSU crystallisation and the clinical presentation of gout [1]. Previous genome-wide association studies (GWAS) have identified 28 single nucleotide polymorphisms (SNPs) associated with serum urate concentration [2]. Some of these SNPs encode renal and/or gut urate transport-related proteins and associate with the risk of gout [2–5]. Others contribute to hyperuricaemia via regulation of purine synthesis and glucose metabolism pathways [2].
Long-established dietary factors for hyperuricaemia and gout risk include red meat, seafood, and alcohol [6, 7]. In addition, multiple studies have reported that increased coffee intake is associated with reduced serum urate [8–12] and risk of developing gout [13, 14]. This is the case for both caffeinated and de-caffeinated coffee. This association has been attributed to several potential mechanisms including improved insulin resistance [15–21] and caffeine-mediated inhibition of xanthine oxidase [8].
SNPs associated with habitual coffee consumption have also been identified by GWAS [22]. Of note, the GWAS of coffee consumption identified a number of genes which overlap with the genes identified in the separate hyperuricaemia GWAS [2, 22]. Specifically, alleles in several genes associated with hyperuricaemia (GCKR, ABCG2, MLXIPL, and CYP1A2 [2]) are also associated with decreased habitual coffee intake [22]. These four alleles are the sole alleles currently known to be associated with both serum urate, gout risk, and habitual coffee consumption from existing genome-wide analysis studies. Regional association plots suggest that the signals for both urate and habitual coffee consumption are very similar for the four loci (Additional file 1: Figure S1). The aim of this study was to determine whether the lead urate-associated SNPs at the four coffee and serum urate associated loci influence the risk of gout through their effects on coffee consumption.
Methods
Participants
This research was conducted using the UK Biobank Resource [23, 24] (approval number 12611). Data for the UK Biobank were gathered over 2006–2010 from people between the ages of 40 and 69 years old. The North West Multi-Centre Research Ethics Committee granted ethical approval for UK Biobank participants. All participants provided written informed consent.
Inclusion criteria for this analysis were European ethnicity (white, British, Irish, any other white background) and genome-wide genotypes available. Exclusion criteria were self-reported sex mismatch with genetic sex, genotyping quality-control failure, and related individuals. Gout cases were ascertained in the UK Biobank using a validated case-definition of “self-reported gout or urate-lowering therapy (ULT) use” [25].
Genotyping and SNP selection
UK Biobank samples were genotyped using an Affymetrix Axiom array (820,967 markers) and imputed to ~ 73.3 M SNPs using SHAPEIT3 and IMPUTE2, with a combined UK10K and 1000 Genomes reference panel.
Details of the lead coffee-associated SNP at each locus reported by Cornelis et al. (2015) [22] were extracted from the publication. The equivalent information was extracted from the serum urate GWAS publication for overlapping loci [2] and aligned to that of the coffee GWAS. For both GCKR and CYP1A2, the lead SNP reported in both studies was identical. For ABCG2 and MLIXPL, the linkage disequilibrium (LD) between the lead urate-associated and coffee-associated SNP was calculated using the European 1000 Genomes phase 3 (September 2014) and UK Biobank data with PLINK v1.90 [26, 27]. The ABCG2 SNPs were in high LD (r21000Genomes = 0.94; r2UKBiobank = 0.99), whilst the MLIXPL SNPs were in moderate LD (r21000Genomes = 0.58; r2UKBiobank = 0.57) (Table 1). Because of the adequate LD, the four urate-associated SNPs, GCKR (rs1260326), ABCG2 (rs2231142), MLXIPL (rs1178977), and CYP1A2 (rs2472297), were employed in the analysis (Table 1).
Table 1.
SNPs associated with serum urate levels and habitual coffee intake in previous GWAS using participants of European ancestry
| GCKR | ABCG2 | MLXIPL | CYP1A2 | ||
|---|---|---|---|---|---|
| Coffee GWAS [20] | SNP | rs1260326 | rs1481012 | rs7800944 | rs2472297 |
| Chr: Position (B37) | 2:27730940 | 4:89039082 | 7:73035857 | 15:75027880 | |
| Effect allele/other | T/C | A/G | T/C | T/C | |
| Effect allele freq. | 0.41 | 0.89 | 0.72 | 0.24 | |
| Beta (cups/day) | −0.04 | 0.06 | −0.05 | 0.15 | |
| SE | 0.01 | 0.01 | 0.01 | 0.01 | |
| P | 1.06 × 10−7 | 1.13 × 10−6 | 7.82 × 10−9 | 6.45 × 10−47 | |
| Urate GWAS [2] | SNP | rs1260326 | rs2231142 | rs1178977 | rs2472297 |
| Chr: Position (B37) | 2:27730940 | 4:89271347 | 7:72494985 | 15:75027880 | |
| Effect allele/other | T/C | G/T | A/G | T/C | |
| Effect allele freq. | 0.41 | 0.89 | 0.81 | 0.24 | |
| Beta (mg/dl) | 0.07 | −0.22 | 0.05 | −0.03 | |
| SE | 0.01 | 0.01 | 0.01 | 0.01 | |
| P | 1.20 × 10−44 | 1.00 × 10−134 | 1.20 × 10−12 | 3.85 × 10−6 | |
| LD | Euro r2 | Same SNP | 0.94 | 0.58 | Same SNP |
| rs2231142: T | rs1178977: G | ||||
| rs1481012: G | rs7800944: C | ||||
| UKBB r2 | Same SNP | 0.99 | 0.57 | Same SNP | |
| rs2231142: T | rs1178977: G | ||||
| rs1481012: G | rs7800944: C |
Linkage disequilibrium calculated using 1000 Genomes phase 3 (September 2014) data
Chr chromosome, Euro European from 1000 Genomes, Freq frequency, GWAS genome-wise association study, LD linkage disequilibrium, SE standard error, SNP single nucleotide polymorphism, UKBB UK Biobank
Coffee and food-frequency intake
At the time of recruitment, data were collected on usual coffee consumption in addition to other foodstuffs [28]. Coffee intake was determined by participants’ answers to the question “How many cups of coffee do you drink each day (include de-caffeinated coffee)?” Values were numerical in exact cups per day (including 0) or under 1 cup per day. Any non-zero value was determined as “any” coffee consumption. Other food intakes were determined by responses to questions about daily or weekly food frequency [28] (Additional file 2: Table S1 for codes).
Statistical analysis
Multivariate linear and logistic regression analysis was completed using SPSS version 24 (IBM, New York, USA). All models were adjusted for age, sex, body mass index (BMI), hypertension, kidney disease, diabetes mellitus, and reported intake of cups of tea, fruit, vegetables, meat, fish, bread, cereal, and cheese. Beer and spirits were included as additional variables in separate analyses due to high levels of missing data, with only 70.4% of participants having information on beer/cider intake and 70.2% of participants with information on spirit intake. The effect of each SNP was analysed using two models, a dominant model (for the presence of least one urate-raising allele) and a recessive model (homozygosity for the urate-raising allele). Experiment-wide significance was set at P < 0.0125 after Bonferroni correction for multiple testing (0.05 divided by four SNPs). For gout, multivariate logistic regression analysis with gout as the dependent variable and coffee intake or SNP status as the independent variables was completed. For coffee intake, multivariate linear regression analysis was completed with coffee intake (cups per day) as the dependent variable and SNP status as the independent variables. All regression analyses were fully adjusted as described above.
The PROCESS macro v2.16.3 for SPSS was used to construct a mediation pathway to gout risk. Model 4 (used for simple mediation models exploring the relationship between a single independent variable, single mediator, and single dependent variable as presented in Additional file 3: Figure S2) was used from the PROCESS macro with 1000 bootstraps. This aimed to quantify the direct and indirect relationships between SNPs, coffee intake, and gout. Only SNPs associated with both gout and coffee consumption in the UK Biobank data (GCKR and ABCG2) were included in the mediation analysis; this aimed to establish whether the effects of GCKR/ABCG2 on gout risk were mediated through coffee intake. Direct and indirect standardised effect estimates were calculated using multivariate-adjusted linear and logistic regression analysis as described above. All effect estimates were adjusted for the same potential confounding variables as described above. Bootstrapping was used to determine whether the indirect effect of the SNP on gout risk through coffee intake was significant.
Results
Participant demographics
Data including dietary information and genome-wide genotypes were available for 130,966 participants, including 2135 participants with gout. The demographic and clinical information of participants is shown in Table 2, with full dietary information shown in Additional file 4 (Table S2). Participants with gout were older, more likely to be male, had higher mean BMI, and had a higher prevalence of co-morbid conditions.
Table 2.
Demographic and clinical characteristics of study population (n = 130,966)
| Controls | % with data available | Gout cases | % with data available | |
|---|---|---|---|---|
| n | 128,831 | 2135 | ||
| Males, n (%) | 59,782 (46.4%) | 100.0 | 1972 (92.4%) | 100.0 |
| Age (years) | 56.7 (7.98) | 100.0 | 60.23 (6.67) | 100.0 |
| Body mass index (kg /m2) | 27.47 (4.82) | 99.7 | 30.87 (5.01) | 99.6 |
| Townsend Index | −1.38 (3.05) | 99.9 | −1.12 (3.10) | 99.8 |
| Hypertension, n (%)) | 40,185 (31.2%) | 100.0 | 1402 (65.7%) | 100,0 |
| Diabetes, n (%) | 5384 (4.2%) | 100.0 | 279 (13.1%) | 100.0 |
| Kidney disease, n (%) | 1121 (0.9%) | 100.0 | 125 (5.9%) | 100.0 |
| Meat consumption, n (%) | 122,658 (95.2%) | 98.9 | 2103 (98.5%) | 99.2 |
| All meat intake (pieces per week) | 5.50 (2.72) | 98.9 | 6.55 (2.77) | 99.2 |
| Fish consumption, n (%) | 123,330 (95.7%) | 99.3 | 2079 (97.4%) | 99.2 |
| All fish intake (pieces per week) | 2.23 (1.58) | 99.3 | 2.34 (1.63) | 99.2 |
| Beer/cider intake (pints per week) | 3.11 (5.71) | 70.2 | 9.11 (10.26) | 83.8 |
| Spirits intake (measures per week) | 1.98 (5.91) | 70.0 | 2.97 (8.99) | 83.4 |
| Coffee consumption, n (%) | 101,076 (78.5%) | 99.8 | 1617 (75.7%) | 99.2 |
| Coffee intake (cups per day) | 2.12 (2.20) | 99.8 | 1.75 (1.83) | 99.2 |
| Tea consumption, n (%) | 108,734 (84.4%) | 99.8 | 1813 (84.9%) | 99.9 |
| Tea intake (cups per day) | 3.45 (2.99) | 99.8 | 3.31 (2.88) | 99.9 |
| Any fruit consumption, n (%) | 119,605 (92.8%) | 98.9 | 1917 (89.8%) | 98.0 |
| Fruit of any description (pieces per day) | 2.98 (2.50) | 98.9 | 2.72 (2.33) | 98.0 |
| All vegetable intake (pieces per day) | 4.83 (3.17) | 98.6 | 4.83 (3.06) | 97.3 |
| Bread intake (slices per week) | 12.52 (8.69) | 99.2 | 14.77 (9.72) | 98.7 |
| Cereal intake (bowls per week) | 4.52 (2.80) | 99.7 | 3.83 (2.81) | 99.7 |
| Cheese intake (pieces per week) | 2.46 (1.75) | 97.7 | 2.35 (1.66) | 96.9 |
Hypertension, diabetes mellitus, and kidney disease defined by self-reported illness or hospital diagnosis
All values are shown as mean (standard deviation) unless otherwise indicated
Coffee consumption and gout
Coffee consumption was inversely associated with gout (Table 3). Multivariate adjusted odds ratio (OR) (95% confidence interval (CI)) of gout for any coffee consumption was 0.75 (0.67–0.84, P = 9 × 10−7) and 0.85 (0.82–0.87, P = 9 × 10−32) per cup of coffee consumed per day. These associations persisted after intake of beer and spirits was added to the models.
Table 3.
Association of coffee intake with gout
| Association of coffee intake (any coffee) with gout | ||||
| Observations | Odds ratio | 95% confidence interval | P | |
| Unadjusted | 130,731 | 0.85 | 0.77–0.94 | 0.002 |
| Adjusted† | 121,897 | 0.75 | 0.67–0.84 | 9.05 × 10−7 |
| Adjusted† including beer/spirits |
86,676 | 0.75 | 0.66–0.86 | 1.40 × 10−5 |
| Association of coffee intake (per cup per day) with gout | ||||
| Observations | Odds ratio | 95% confidence Interval | P | |
| Unadjusted | 130,731 | 0.91 | 0.89–0.93 | 3.08 × 10−15 |
| Adjusted† | 121,897 | 0.85 | 0.82–0.87 | 8.55 × 10−32 |
| Adjusted† including beer/spirits |
86,676 | 0.85 | 0.83–0.88 | 1.09 × 10−23 |
†Adjusted for age, sex, body mass index, hypertension, kidney disease, diabetes, meat intake, fish intake, cheese intake, tea intake, fruit intake, vegetable intake, bread intake, and cereal intake
Association of SNPs with gout risk
The urate-associated SNPs for both GCKR and ABCG2 were significantly associated with gout (Table 4). The strongest observed association was for ABCG2 with multivariate adjusted OR 2.37 (95% CI 2.15–2.61, P = 6 × 10−69). GCKR also had a strong effect on gout risk with multivariate adjusted OR 1.43 (95% CI 1.29–1.58, P = 4 × 10−12). Neither MLXIPL nor CYP1A2 were associated with gout, with multivariate adjusted OR 1.24 (95% CI 0.95–1.61, P = 0.11) and 1.16 (95% CI 0.96–1.40, P = 0.12), respectively. For GCKR and ABCG2, association with gout was observed for both the presence of at least one urate-raising allele (Table 4) as well as by number of urate-raising alleles (Additional file 5: Table S3).
Table 4.
Association analysis of urate-associated SNPs with gout and coffee intake
| Association of gout with urate-associated SNPs | Association of coffee intake (cups per day) with urate-associated SNPs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Effect allele | Observations | % | Odds ratio | 95% Confidence interval | P | Observations | % | Unadjusted β coefficient | 95% Confidence interval |
P |
| GCKR | ||||||||||||
| Unadjusted | rs1260326 | T | 130,966 | 100.0% | 1.40 | 1.27–1.53 | 3.34 × 10−12 | 130,731 | 100.0% | −0.07 | −0.10 to −0.05 | 1.33 × 10−8 |
| Adjusted† | 121,940 | 93.1% | 1.43 | 1.29–1.58 | 4.05 × 10−12 | 121,897 | 93.2% | −0.08 | −0.10 to −0.05 | 3.05 × 10−10 | ||
| Adjusted† including beer/spirits | 86,702 | 66.2% | 1.45 | 1.30–1.62 | 2.32 × 10−11 | 86,676 | 66.3% | −0.07 | −0.10 to −0.05 | 2.47 × 10−8 | ||
| ABCG2 | ||||||||||||
| Unadjusted | rs2231142 | T | 130,966 | 100.0% | 2.26 | 2.07–2.47 | 1.05 × 10−72 | 130,731 | 100.0% | −0.07 | −0.10 to −0.04 | 1.00 × 10−6 |
| Adjusted† | 121,940 | 93.1% | 2.37 | 2.15–2.61 | 6.62 × 10−69 | 121,897 | 93.2% | −0.09 | −0.11 to −0.06 | 2.10 × 10−9 | ||
| Adjusted† including beer/spirits | 86,702 | 66.2% | 2.45 | 2.20–2.72 | 4.78 × 10−61 | 86,676 | 66.3% | −0.08 | −0.11 to −0.05 | 3.99 × 10−7 | ||
| MLXIPL | ||||||||||||
| Unadjusted | rs1178977 | A | 130,966 | 100.0% | 1.25 | 0.98–1.61 | 0.07 | 130,702 | 100.0% | −0.15 | −0.21 to −0.09 | 3.00 × 10−6 |
| Adjusted† | 121,940 | 93.1% | 1.24 | 0.95–1.61 | 0.11 | 121,897 | 93.2% | −0.17 | −0.23 to −0.11 | 3.20 × 10−8 | ||
| Adjusted† including beer/spirits | 86,702 | 66.2% | 1.24 | 0.94–1.65 | 0.13 | 86,661 | 66.3% | −0.21 | −0.27 to −0.14 | 5.45 × 10−10 | ||
| CYP1A2 | ||||||||||||
| Unadjusted | rs2472297 | C | 130,966 | 100.0% | 1.15 | 0.96–1.37 | 0.13 | 130,731 | 100.0% | −0.25 | −0.30 to −0.21 | 3.02 × 10−26 |
| Adjusted† | 121,940 | 93.1% | 1.16 | 0.96–1.40 | 0.12 | 121,897 | 93.2% | −0.30 | −0.35 to −0.26 | 7.61 × 10−40 | ||
| Adjusted† including beer/spirits | 86,702 | 66.2% | 1.16 | 0.94–1.42 | 0.17 | 86,676 | 66.3% | −0.30 | −0.35 to −0.25 | 2.80 × 10−32 | ||
Analysis is shown for the presence of at least one urate-raising allele
Effect allele is allele associated with hyperuricaemia in Kottgen GWAS paper [2]
SNP single nucleotide polymorphism
†Adjusted for age, sex, body mass index, hypertension, kidney disease, diabetes, meat intake, fish intake, cheese intake, tea intake, fruit intake, vegetable intake, bread intake, and cereal intake
Association of SNPs with coffee intake
All four urate-associated SNPs were also associated with reduced coffee intake (Table 4). CYP1A2 showed the strongest association with reduced coffee intake (beta −0.30, 95% CI −0.35 to −0.26, P = 8 × 10−40), followed by MLXIPL (beta −0.17, 95% CI −0.23 to −0.11, P = 3 × 10−8). Weaker associations were observed with GCKR (beta −0.08, 95% CI −0.11 to −0.06, P = 3 × 10−10) and ABCG2 (beta −0.09, 95% CI −0.11 to −0.06, P = 2 × 10−9). All SNPs demonstrated evidence of a dose effect for coffee consumption with the strongest association evident in those with two copies of the urate-raising allele (Additional file 6: Table S4).
Mediation analysis
In logistic regression models that included urate- and gout-associated SNPs, coffee consumption was inversely associated with gout (Table 5). In these models, the association of both GCKR and ABCG2 with gout were also observed. Association of coffee consumption with gout was independent of all SNPs, and association of GCKR and ABCG2 SNPs with gout was independent of coffee consumption.
Table 5.
Logistic regression models including adjustment for both coffee intake and urate-associated SNPs for association with gout
| Variable | Odds ratio for gout | 95% Confidence Interval | Standard Error | P |
|---|---|---|---|---|
| GCKR (rs1260326) | 1.42 | 1.28 - 1.57 | 0.05 | 7.78E-12 |
| Coffee consumption (any) | 0.76 | 0.68 - 0.85 | 0.06 | 2.00E-06 |
| ABCG2 (rs2231142) | 2.36 | 2.14 -2.60 | 0.05 | 2.07E-68 |
| Coffee consumption (any) | 0.76 | 0.68 - 0.85 | 0.06 | 2.00E-06 |
| MLXIPL (rs1178977) | 1.23 | 0.95 -1.60 | 0.13 | 0.12 |
| Coffee consumption (any) | 0.75 | 0.67 - 0.84 | 0.06 | 9.45E-07 |
| CYP1A2 (rs2472297) | 1.15 | 0.96 - 1.39 | 0.10 | 0.14 |
| Coffee consumption (any) | 0.75 | 0.67 - 0.84 | 0.06 | 1.00E-06 |
| GCKR (rs1260326) | 1.41 | 1.27 - 1.55 | 0.05 | 2.79E-11 |
| Coffee intake (per cup per day) | 0.85 | 0.83 - 0.87 | 0.01 | 4.29E-31 |
| ABCG2 (rs2231142) | 2.33 | 2.12 - 2.57 | 0.05 | 1.83E-66 |
| Coffee intake (per cup per day) | 0.85 | 0.83 - 0.88 | 0.01 | 4.02E-30 |
| MLXIPL (rs1178977) | 1.20 | 0.92 - 1.56 | 0.13 | 0.17 |
| Coffee intake (per cup per day) | 0.85 | 0.82 - 0.87 | 0.01 | 1.15E-31 |
| CYP1A2 (rs2472297) | 1.11 | 0.92 - 1.34 | 0.10 | 0.27 |
| Coffee intake (per cup per day) | 0.85 | 0.83 - 0.87 | 0.01 | 1.41E-31 |
Effect allele is allele associated with hyperuricaemia in Kottgen GWAS paper (2)
Odds are shown for presence/absence of urate-raising allele
All results shown are adjusted for age, sex, body mass index, hypertension, kidney disease, diabetes, meat intake, fish intake, cheese intake, tea intake, fruit intake, vegetable intake, bread intake, and cereal intake
SNP single nucleotide polymorphism
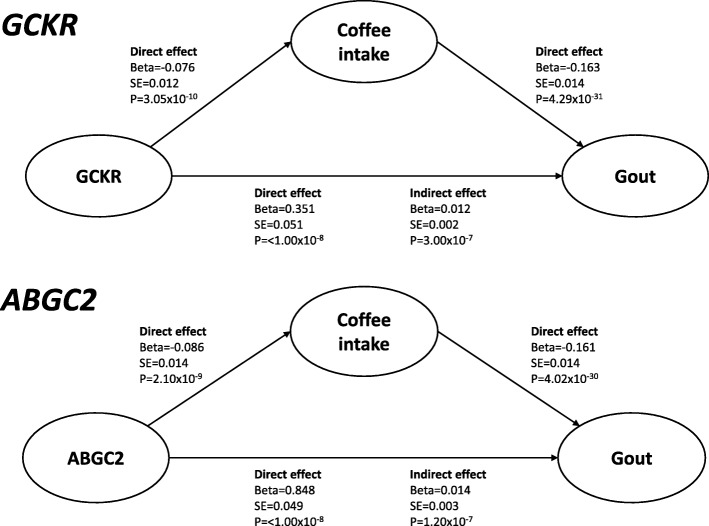
Mediation results are shown in Fig. 1. In mediation analysis of GCKR, a strong direct effect of the urate-associated SNP on gout risk was demonstrated (beta 0.351, standard error (SE) 0.051, P < 1.00 × 10−8). A direct effect of the urate-associated SNP on coffee intake was also demonstrated (beta −0.076, SE = 0.012, P = 3.05 × 10−10). The direct effect of the urate-associated SNP accounted for most of the total observed effect on gout risk, with much smaller indirect effects mediated by coffee consumption (beta 0.012, SE 0.002 compared with beta 0.351, SE 0.051).
Fig. 1.
Summary of mediation analysis. Standardised path coefficients are shown. The direction of the path analysis from SNP to gout was pre-specified. Analysis is shown for the presence of at least one urate-raising allele and coffee intake in cups per day. Effect sizes are shown when results are adjusted for the following co-variates: age, sex, body mass index, hypertension, kidney disease, diabetes mellitus, and intake of meat, fish, cheese, tea, fruit, vegetables, bread, and cereal. SE standard error
Similarly, in mediation analysis of ABCG2, a strong direct effect of the urate-associated SNP on gout risk was demonstrated (beta 0.848, SE 0.049, P < 1.00 × 10−8). A direct effect of the urate-associated SNP on coffee intake was also demonstrated (beta −0.086, SE 0.014, P = 2.10 × 10−9). The direct effect of the urate-associated ABCG2 SNP accounted for most of the total observed effect on gout risk, with a much smaller indirect effect mediated by coffee consumption (beta 0.848, SE 0.049 compared with beta 0.014, SE 0.03).
Discussion
This study provides further evidence that coffee intake is inversely associated with gout. This association was observed in those who consumed any coffee versus none, with evidence of a dose-effect response for the number of cups consumed per day. Although the urate-raising GCKR and ABCG2 alleles were associated with both lower coffee consumption and higher risk of gout, mediation analysis demonstrated that these SNPs largely influence gout risk directly, rather than indirectly through their dual effect on coffee consumption.
In our analysis of the UK Biobank, all of the loci from previous GWAS of habitual coffee consumption [22] tested here (ABCG2, GKCR, MLXIPL, and CYP1A2) were replicated, strengthening evidence for the genetic basis of this trait. CYP1A2 and its product, cytochrome P450, have been demonstrated to have a central role in the metabolism of caffeine [29]. Indeed, CYP1A2 is one of the strongest known loci for coffee consumption, both in this report and in other studies [30]. Our results are consistent with a recent GWAS meta-analysis that reported genome-wide significant associations of the CYP1A2 locus and nominal association of the GCKR and ABCG2 loci with plasma caffeine levels [31].
Despite the association of GCKR and ABCG2 with both gout and reduced coffee intake, our mediation analysis indicates that the dominant mechanism for GCKR and ABCG2 on gout risk is not through coffee consumption. The urate-raising alleles of these SNPs are associated with reduced coffee intake; this suggests that implementing a dietary intervention such as increased coffee consumption may be more difficult in those genetically pre-disposed towards gout as urate levels would be raised. Analysis of modifiable exposures associated with gout risk alleles is an avenue for further study as GWAS for various dietary factors become more common. Indeed, a recent UK Biobank study identified several SNPs associated with alcohol consumption, including GCKR rs1260326 [32]. This is especially interesting given the recently reported interaction between GCKR rs780094 (in very strong linkage disequilibrium with rs1260326), alcohol, and gout risk [33].
We did not use Mendelian randomisation to directly address the question of the causality of coffee consumption in regulating serum urate levels and the risk of gout. This is because the majority of the coffee consumption-associated genetic variants also associate with serum urate levels making the variants unsuitable as Mendelian randomisation instrumental variables. However, we note that the AHR (aryl hydrocarbon receptor) locus, with an approximately equal effect size on coffee consumption as CYP1A2 [22], does not associate with serum urate levels [2]. Consistent with our mediation analysis, this does not support a direct causal role of coffee consumption in regulating serum urate levels. Very recently, a conventional Mendelian randomisation study [34] provided support for a causal role of coffee consumption in reducing the risk of gout. However, the authors of this Mendelian randomisation also observed in the separate gout case-control data set [2] that genetic variation in AHR, one of the strongest genetic effects on habitual coffee consumption [22], did not contribute to the protective effect for gout. One of the fundamental assumptions of Mendelian randomisation is that the SNPs used for the instrumental variables are unrelated to the outcome [35]. Although statistical methods exist for evaluating the effect of pleiotropy in Mendelian randomisation studies, the fact that coffee- and urate-associated loci overlap limits the application of Mendelian randomisation to assessing a possible causal role of coffee in gout.
This study has several limitations. Only three of the four urate-associated SNPs used in this study were identical to (or in very high LD with) the SNPs reported in the previous coffee GWAS. However, we did observe an association between coffee consumption and all four urate-associated SNPs tested in this analysis. The study population is restricted to those of white European ancestry, thereby limiting the generalisability to other populations. There may also be some bias in reported dietary intakes. Furthermore, widely used food-frequency questionnaires lack specificity for some foodstuffs [36]. Any effect of this is likely minimised due to the UK Biobank’s extremely comprehensive data collection gathered through multiple modalities. Similarly, the non-specific nature of data collection for general use rather than specific research questions may also help minimise the effects of recall bias. Key strengths of this study include the large sample size and wide access to multiple data sources such as self-reported illness, medications, hospital records, and dietary data.
Conclusions
In summary, this work further supports the hypothesis that coffee consumption is protective for gout risk. The exact mechanism of this protective effect remains unclear. Although several SNPs associate with both lower total coffee consumption and higher gout risk, mediation analysis indicates that these SNPs have direct effects on gout risk rather than indirect effects mediated by coffee consumption. The coffee- and urate-associated loci could influence coffee consumption and urate levels, respectively, through separate biological mechanisms.
Additional files
Figure S1. Regional association plots of genome-wide significant urate and habitual coffee loci. In each panel, SNPs identified as associated with both urate [2] and coffee intake [22] are plotted with their –log10 (P values) as a function of genomic position using HG build 19 and 1000 genomes European reference for LD (November 2014). Each SNP is coloured according to its correlation with the index SNP (demonstrating the lowest P value within the region, labelled in purple) according to a scale from r2 = 0 to r2 = 1. Urate-raising alleles are displayed on the left and coffee-associated alleles are on the right. LocusZoom plots were drawn from publicly available data ex [2] and taken from [22]. GCKR rs2911711 is in complete linkage disequilibrium with rs1260326. (PDF 826 kb)
Table S1. Coding of food-frequency intakes for analysis. (DOC 40 kb)
Figure S2. Description of PROCESS model 4. (PDF 29 kb)
Table S2. Complete dietary information for the study population. (DOCX 16 kb)
Table S3. Association analysis of genotype with gout risk. (DOC 57 kb)
Table S4. Association analysis of genotype with habitual coffee intake. (DOC 54 kb)
Acknowledgements
This work was conducted using the UK Biobank Resource (approval number 12611). We thank Dr. Marilyn Cornelis and the Coffee and Caffeine Genetics Consortium for sharing regional association data to generate LocusZoom plot files for Additional file 1: Figure S1.
Funding
This work was supported by the Health Research Council of New Zealand (grant number 14–527).
Availability of data and materials
The data in this study is owned by a third party, UK Biobank (www.ukbiobank.ac.uk). Legal constrains do not permit public data sharing but is open to all bona fide researchers worldwide. The data reported can therefore be directly accessed by applying through the UK Biobank Access Management System (www.ukbiobank.ac.uk/register-apply).
Abbreviations
- ABCG2
ATP-binding cassette subfamily G member 2
- BMI
Body mass index
- CI
Confidence interval
- CYP1A2
Cytochrome P450 family 1 subfamily A member 2
- GCKR
Glucokinase regulator
- GWAS
Genome-wide association studies
- LD
Linkage disequilibrium
- MLXIPL
MAX-like protein X interacting protein like
- MSU
Monosodium urate
- OR
Odds ratio
- SE
Standard error
- SNP
Single nucleotide polymorphism
- ULT
Urate-lowering therapy
Authors’ contributions
JH analysed the data and drafted the manuscript. TRM and ND contributed to study design, data analysis, data interpretation, and contributed to the manuscript draft. RT obtained the dataset from the UK Biobank and assisted with producing the Locus Zooms plots. TJM analysed the overlapping SNPs in both Kottgen and Cornwallis datasets. TF assisted with producing the Locus Zoom plots. All authors read and approved the final manuscript. All authors are accountable for all aspects of the work.
Ethics approval and consent to participate
UK Biobank obtained approval from the North West Multi-Centre Research Ethics Committee (11/NW/0382). Full written informed consent was obtained from all participants prior to the study.
Competing interests
TRM has received consulting fees or grants from Ardea Biosciences and AstraZeneca. ND has received consulting fees, speaker fees, or grants from Takeda, Teijin, Menarini, Pfizer, Ardea Biosciences, AstraZeneca, Fonterra, Crealta, and Cymabay. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Joseph Hutton, Tony R. Merriman and Nicola Dalbeth contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s13075-018-1629-5) contains supplementary material, which is available to authorized users.
Contributor Information
Joseph Hutton, Email: jh7480@my.bristol.ac.uk.
Tahzeeb Fatima, Email: tahzeeb.fatima@otago.ac.nz.
Tanya J. Major, Email: tanya.major@otago.ac.nz
Ruth Topless, Email: ruth.topless@otago.ac.nz.
Lisa K. Stamp, Email: lisa.stamp@cdhb.health.nz
Tony R. Merriman, Email: tony.merriman@otago.ac.nz
Nicola Dalbeth, Email: n.dalbeth@auckland.ac.nz.
References
- 1.Dalbeth N, Merriman TR, Stamp LK. Gout. Lancet. 2016;388(10055):2039–2052. doi: 10.1016/S0140-6736(16)00346-9. [DOI] [PubMed] [Google Scholar]
- 2.Kottgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45(2):145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration. urate excretion and gout Nature genetics. 2008;40(4):437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 4.Woodward OM, Kottgen A, Coresh J, Boerwinkle E, Guggino WB, Kottgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. P Natl Acad Sci USA. 2009;106(25):10338–10342. doi: 10.1073/pnas.0901249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merriman TR. An update on the genetic architecture of hyperuricemia and gout. Arthritis research & therapy. 2015;17:98. doi: 10.1186/s13075-015-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi HK, Atkinson K, Karlson EW, Willett W, Curhan G. Purine-rich foods, dairy and protein intake, and the risk of gout in men. N Engl J Med. 2004;350(11):1093–1103. doi: 10.1056/NEJMoa035700. [DOI] [PubMed] [Google Scholar]
- 7.Choi HK, Atkinson K, Karlson EW, Willett W, Curhan G. Alcohol intake and risk of incident gout in men: a prospective study. Lancet. 2004;363(9417):1277–1281. doi: 10.1016/S0140-6736(04)16000-5. [DOI] [PubMed] [Google Scholar]
- 8.Kela U, Vijayvargiya R, Trivedi CP. Inhibitory effects of methylxanthines on the activity of xanthine-oxidase. Life Sci. 1980;27(22):2109–2119. doi: 10.1016/0024-3205(80)90492-0. [DOI] [PubMed] [Google Scholar]
- 9.Kiyohara C, Kono S, Honjo S, Todoroki I, Sakurai Y, Nishiwaki M, et al. Inverse association between coffee drinking and serum uric acid concentrations in middle-aged Japanese males. Brit. J Nutr. 1999;82(2):125–130. [PubMed] [Google Scholar]
- 10.Choi HK, Curhan G. Coffee, tea, and caffeine consumption and serum uric acid level: the third National Health and nutrition examination survey. Arthrit Rheum-Arthr. 2007;57(5):816–821. doi: 10.1002/art.22762. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Yang T, Zeng C, Wei J, Li H, Xiong YL, et al. Is coffee consumption associated with a lower risk of hyperuricaemia or gout? A systematic review and meta-analysis. BMJ Open. 2016;6(7):e009809. doi: 10.1136/bmjopen-2015-009809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park KY, Kim HJ, Ahn HS, Kim SH, Park EJ, Yim SY, et al. Effects of coffee consumption on serum uric acid: systematic review and meta-analysis. Semin Arthritis Rheum. 2016;45(5):580–586. doi: 10.1016/j.semarthrit.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Choi HK, Willett W, Curhan G. Coffee consumption and risk of incident gout in men—a prospective study. Arthritis Rheum. 2007;56(6):2049–2055. doi: 10.1002/art.22712. [DOI] [PubMed] [Google Scholar]
- 14.Choi HK, Curhan G. Coffee consumption and risk of incident gout in women: the Nurses’ health study. Am J Clin Nutr. 2010;92(4):922–927. doi: 10.3945/ajcn.2010.29565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salazar-Martinez E, Willett WC, Ascherio A, Manson JE, Leitzmann MF, Stampfer MJ, et al. Coffee consumption and risk for type 2 diabetes mellitus. Ann Intern Med. 2004;140(1):1–8. doi: 10.7326/0003-4819-140-1-200401060-00005. [DOI] [PubMed] [Google Scholar]
- 16.Greer F, Hudson R, Ross R, Graham T. Caffeine ingestion decreases glucose disposal during a hyperinsulinemic-euglycemic clamp in sedentary humans. Diabetes. 2001;50(10):2349–2354. doi: 10.2337/diabetes.50.10.2349. [DOI] [PubMed] [Google Scholar]
- 17.Keijzers GB, De Galan BE, Tack CJ, Smits P. Caffeine can decrease insulin sensitivity in humans. Diabetes Care. 2002;25(2):364–369. doi: 10.2337/diacare.25.2.364. [DOI] [PubMed] [Google Scholar]
- 18.Thong FSL, Graham TE. Caffeine-induced impairment of glucose tolerance is abolished by beta-adrenergic receptor blockade in humans. J Appl Physiol. 2002;92(6):2347–2352. doi: 10.1152/japplphysiol.01229.2001. [DOI] [PubMed] [Google Scholar]
- 19.van Dam RM, Hu FB. Coffee consumption and risk of type 2 diabetes—a systematic review. J Am Med Assoc. 2005;294(1):97–104. doi: 10.1001/jama.294.1.97. [DOI] [PubMed] [Google Scholar]
- 20.Petrie HJ, Chown SE, Belfie LM, Duncan AM, McLaren DH, Conquer JA, et al. Caffeine ingestion increases the insulin response to an oral-glucose-tolerance test in obese men before and after weight loss. Am J Clin Nutr. 2004;80(1):22–28. doi: 10.1093/ajcn/80.1.22. [DOI] [PubMed] [Google Scholar]
- 21.Wu TY, Hankinson SE, Willett WC, Giovannucci E. Caffeinated coffee, decaffeinated coffee, and caffeine in relation to plasma C-peptide levels, a marker of insulin secretion, in US women. Diabetes Care. 2005;28(6):1390–1396. doi: 10.2337/diacare.28.6.1390. [DOI] [PubMed] [Google Scholar]
- 22.Cornelis MC, Byrne EM, Esko T, Nalls MA, Ganna A, Paynter N, et al. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol Psychiatry. 2015;20(5):647–656. doi: 10.1038/mp.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ollier W, Sprosen T, Peakman T. UK biobank: from concept to reality. Pharmacogenomics. 2005;6(6):639–646. doi: 10.2217/14622416.6.6.639. [DOI] [PubMed] [Google Scholar]
- 24.Collins R. Manchester: UK biobank coordinating Centre. 2007. UK biobank: protocol for a large-scale prospective epidemiological resource. [Google Scholar]
- 25.Cadzow M, Merriman TR, Dalbeth N. Performance of gout definitions for genetic epidemiological studies: analysis of UK biobank. Arthritis Res Ther. 2017;19(1):181. doi: 10.1186/s13075-017-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UK Biobank touch-screen questionnaire: final version. UK: Biobank Coordinating Centre; 2006. Available from: http://www.ukbiobank.ac.uk/wp-content/uploads/2011/06/Touch_screen_questionnaire.pdf.
- 29.Perera V, Gross AS, McLachlan AJ. Measurement of CYP1A2 activity: a focus on caffeine as a probe. Curr Drug Metab. 2012;13(5):667–678. doi: 10.2174/1389200211209050667. [DOI] [PubMed] [Google Scholar]
- 30.Thorn CF, Aklillu E, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for CYP1A2. Pharmacogenet Genomics. 2012;22(1):73–77. doi: 10.1097/FPC.0b013e32834c6efd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornelis MC, Kacprowski T, Menni C, Gustafsson S, Pivin E, Adamski J, et al. Genome-wide association study of caffeine metabolites provides new insights to caffeine metabolism and dietary caffeine-consumption behavior. Hum Mol Genet. 2016;25(24):5472–5482. doi: 10.1093/hmg/ddw334. [DOI] [PubMed] [Google Scholar]
- 32.Clarke TKAM, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray A, Smith B, Campbell A, Hayward C, Porteous D, Deary IJ, McIntosh AM. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK biobank (N=112,117) 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasheed H, Stamp LK, Dalbeth N, Merriman TR. Interaction of the GCKR and A1CF loci with alcohol consumption to influence the risk of gout. Arthritis Res Ther. 2017;19(1):161. doi: 10.1186/s13075-017-1369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson SC, Carlstrom M. Coffee consumption and gout: a Mendelian randomisation study. Ann Rheum Dis. 2018. 10.1136/annrheumdis-2018-213055. [Epub ahead of print]. [DOI] [PubMed]
- 35.Robinson PC, Choi HK, Do R, Merriman TR. Insight into rheumatological cause and effect through the use of Mendelian randomization. Nat Rev Rheumatol. 2017;13(3):193. doi: 10.1038/nrrheum.2017.18. [DOI] [PubMed] [Google Scholar]
- 36.Galante J, Adamska L, Young A, Young H, Littlejohns TJ, Gallacher J, et al. The acceptability of repeat internet-based hybrid diet assessment of previous 24-h dietary intake: administration of the Oxford WebQ in UK biobank. Br J Nutr. 2016;115(4):681–686. doi: 10.1017/S0007114515004821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Regional association plots of genome-wide significant urate and habitual coffee loci. In each panel, SNPs identified as associated with both urate [2] and coffee intake [22] are plotted with their –log10 (P values) as a function of genomic position using HG build 19 and 1000 genomes European reference for LD (November 2014). Each SNP is coloured according to its correlation with the index SNP (demonstrating the lowest P value within the region, labelled in purple) according to a scale from r2 = 0 to r2 = 1. Urate-raising alleles are displayed on the left and coffee-associated alleles are on the right. LocusZoom plots were drawn from publicly available data ex [2] and taken from [22]. GCKR rs2911711 is in complete linkage disequilibrium with rs1260326. (PDF 826 kb)
Table S1. Coding of food-frequency intakes for analysis. (DOC 40 kb)
Figure S2. Description of PROCESS model 4. (PDF 29 kb)
Table S2. Complete dietary information for the study population. (DOCX 16 kb)
Table S3. Association analysis of genotype with gout risk. (DOC 57 kb)
Table S4. Association analysis of genotype with habitual coffee intake. (DOC 54 kb)
Data Availability Statement
The data in this study is owned by a third party, UK Biobank (www.ukbiobank.ac.uk). Legal constrains do not permit public data sharing but is open to all bona fide researchers worldwide. The data reported can therefore be directly accessed by applying through the UK Biobank Access Management System (www.ukbiobank.ac.uk/register-apply).