Abstract
Current large-scale recombinant adeno-associated virus (rAAV) production systems based on the baculovirus expression vector (BEV) remain complicated and cost-intensive, and they lack versatility and flexibility. Here we present a novel recombinant baculovirus integrated with all packaging elements for the production of rAAV. To optimize BEV construction, ribosome leaky-scanning mechanism was used to express AAV Rep and Cap proteins downstream of the PH and P10 promoters in the pFast.Bac.Dual vector, respectively, and the rAAV genome was inserted between the two promoters. The yields of rAAV2, rAAV8, and rAAV9 derived from the BEV-infected Sf9 cells exceeded 105 vector genomes (VG) per cell. The BEV was shown to be stable and showed no apparent decrease of rAAV yield after at least four serial passages. The rAAVs derived from the new Bac system displayed high-quality and high-transduction activity. Additionally, rAAV2 could be efficiently generated from BEV-infected beet armyworm larvae at a per-larvae yield of 2.75 ± 1.66 × 1010 VG. The rAAV2 derived from larvae showed a structure similar to the rAAV2 derived from HEK293 cells, and it also displayed high-transduction activity. In summary, the novel BEV is ideally suitable for large-scale rAAV production. Further, this study exploits a potential cost-efficient platform for rAAV production in insect larvae.
Keywords: recombinant baculovirus, rAAV production, insect cells and larvae, gene therapy
Introduction
Recombinant adeno-associated virus (rAAV) is a promising viral vector for gene transfer in both basic research and clinical gene therapy applications.1, 2 In recent years, there has been an increasing need for rAAVs for large animal studies and human clinical trials.3, 4, 5 Many clinical trials using rAAV vectors have had positive outcomes such as in the treatments of monogenic disorders, including Leber congenital amaurosis,6 choroideremia,7 and hemophilia B.8 In 2012, the first rAAV-based drug Glyber was approved in the European Union for the treatment of lipoprotein lipase (LPL) deficiency.9 However, the very high cost of this drug means that many patients cannot afford access to it. As such, the cost-efficient large-scale production of rAAV, while keeping high yield and high quality, remains a major obstacle for rAAV clinical applications.10
The genome of the rAAV vector includes the exogenous gene of interest (GOI) flanked by inverted terminal repeats (ITRs), with the AAV Rep and Cap proteins being supplied in trans.11 Traditional plasmid transfection-based methods are difficult for scale-up production of rAAV, so alternatives must be considered.12, 13 Although adenovirus (ADV) or herpes simplex virus (HSV) infection-based systems have developed for large-scale rAAV production, the downstream purification is hampered by existing pathogenic ADV or HSV.14, 15, 16 Among current large-scale rAAV production systems, the baculovirus expression vector (BEV)-mediated systems have unique advantages, such as high infection efficiency, the non-pathogenicity of BEV, and the cost-efficiency of serum-free Sf9 cell suspension culture.12, 17 In 2002, Urabe et al. established the first-generation Bac system for the production of rAAV.18 In this system, Sf9 cells are co-infected with three BEVs: BEV/Rep, BEV/Cap, and BEV/(ITR-GOI). Because of the genetic instability of BEV/Rep and low co-infection ratio, this three-Bac system is not widely used.19
To improve the stability of BEV/Rep, two strategies have been developed to optimize the Rep expression cassette. In 2008, Chen developed a system to express AAV Rep and Cap proteins from a promoter within an artificial intron.20 In 2009, Smith et al. modified the AAV Rep gene to encode a single bi-functional mRNA transcript that could be translated into two distinct polypeptides, Rep78 and Rep52, by ribosome leaky scanning; meanwhile, the Cap gene was optimized to enhance VP1 protein translation through an optimal Kozak sequence and initiation codon.21 Thus, BEV/Cap-Rep was stable and showed no apparent loss of Cap and Rep protein expression even after five or seven consecutive amplifications. To overcome the drawback of low ratio with multi-BEV co-infection, Aslanidi et al. developed an inducible Sf9/Cap-Rep packaging cell line-based OneBac system in 2009. This strategy improves rAAV yield 10-fold compared with previously described Bac systems.22 In 2014, Mietzsch et al. used the OneBac system to produce the serotypes rAAV1 through rAAV12, and they showed that the rAAV yield reaches about 5 × 105 vector genomes per cell (VG/cell).23 Recently, Mietzsch et al. further developed an improved OneBac2.0 system, which can produce AAV5 with enhanced infectivity and AAV1, AAV2, AAV5, and AAV8 with minimal encapsidation of foreign DNA.24, 25 However, even current packaging cell line-based OneBac systems still have some disadvantages, such as lack of versatility and flexibility, and it is difficult to obtain highly producing Sf9/Cap-Rep packaging cell line in the screenning process.
In this study, we used an optimal strategy to integrate all rAAV-packaging elements, including the Rep gene, the Cap gene, and the rAAV genome (ITR-GOI), in a single BEV without disturbing one another’s functions. We show that the yields of rAAV2, rAAV8, and rAAV9 derived from the new Bac system exceeded 105 VG/cell. We further show that the BEV/Cap-(ITR-GOI)-Rep is stable and showed no apparent decrease of rAAV yield after four serial amplification passages. We therefore posit that the new Bac system is versatile and flexible for large-scale rAAV production. Further, we demonstrate success in producing rAAV2 in beet armyworm larvae infected with the new BEV. These larvae-derived rAAV2 showed a structure similar to the rAAV2 derived from HEK293 cells and high-transduction activity, with an average rAAV2 yield exceeding 1010 VG per larvae. The insect larvae could, therefore, be a potential cost-efficient platform for rAAV production.
Results
Generation of rAAVs from Single Novel BEV-Infected Sf9 Cells
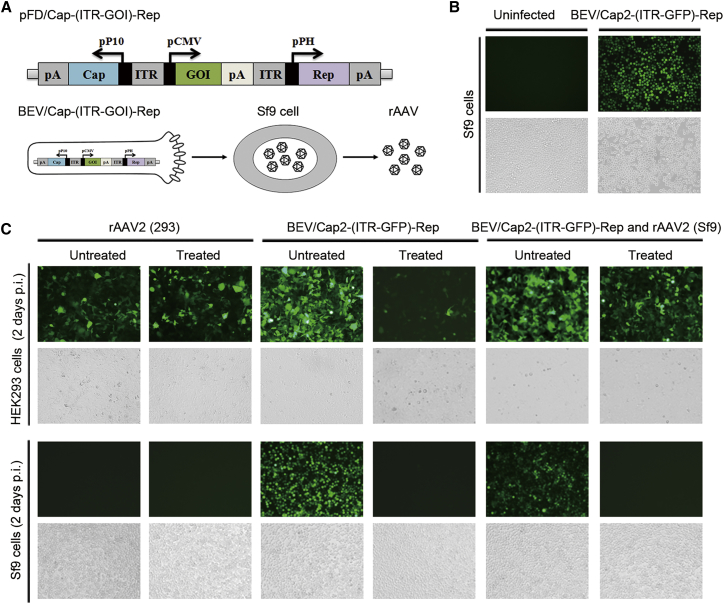
Current BEV-mediated production systems of rAAV have usually been based on Sf9 cells and recombinant baculovirus derived from Autographa californica multiple nucleopolyhedrovirus (AcMNPV).26, 27 In these Bac systems, the rAAV genome (ITR-GOI) is usually integrated in a single BEV, while the AAV Cap and Rep helper genes are supplied by other BEVs or Sf9 packaging cell lines.18, 20, 21, 22 As of yet, there are no reports of a rAAV produced with a single BEV delivering all rAAV-packaging elements. As such, it is a huge challenge to combine all rAAV-packaging elements in a single BEV without functional disruptions. To achieve this goal, we have optimized the organization of the rAAV-packaging elements. The ribosome leaky-scanning mechanism was used to express AAV Rep and Cap proteins, as previously described.21 We chose the AccIII enzyme cut site to insert the rAAV genome (ITR-GOI) between the PH and P10 promoter regions of the pFast.Bac.Dual (pFD) plasmid. To facilitate rAAV detection, the rAAV genome used the cytomegalovirus (CMV) promoter-driven GFP reporter gene expression cassette flanked by AAV2 ITRs. Finally, we constructed the pFD/Cap-(ITR-GFP)-Rep shuttle vector (Figure 1A). Representative experiments used typical rAAV2 for functional validation.
Figure 1.
Schematic of the New Bac System and Verification for rAAV Production upon Single BEV/Cap-(ITR-GFP)-Rep Infection of Sf9 Cells
(A) Schematic of pFD/Cap-(ITR-GFP)-Rep construct and rAAV production upon BEV/Cap-(ITR-GFP)-Rep infection of Sf9 cells. The functional elements are represented by open boxes and separated by vertical lines. Promoters are represented by arrows. (B) Fluorescence microscopy observation of GFP expression in BEV/Cap2-(ITR-GFP)-Rep-infected Sf9 Cells. (C) Fluorescence microscopy observation of 96-well plate cultured HEK293 and Sf9 cells infected with HEK293 cell-derived AAV2 (293) at an MOI of 5,000, supernatants of BEV/Cap2-(ITR-GFP)-Rep-infected Sf9 cells (10 μL culture supernatants per well), and supernatants of BEV/Cap2-(ITR-GFP)-Rep-infected Sf9 cells lysate samples (10 μL lysate supernatants of 1 × 104 cells Sf9 cells per well), respectively. Treated, heat treatment with 60°C for 30 min; p.i., post-infection. Representative fields are shown; upper images are green fluorescence fields, lower images are bright fields.
To obtain the BEVs, the pFD/Cap-(ITR-GFP)-Rep plasmids were transformed into the DH10Bac strain, and then the positive Bacmid DNA was transfected into Sf9 cells. By 72 hr post-transfection, the transfected Sf9 cells showed apparent GFP expression and cytopathic effect (Figure 1B). The data indicate that the BEV/Cap2-(ITR-GFP)-Rep was successfully generated. The culture supernatants of the transfected Sf9 cells were collected as BEV stock P1. We then used the P1 to infect naive Sf9 cells at an MOI of 3. After 72 hr post-infection, we collected the culture supernatants and harvested the cells.
The differences in thermo-stability between enveloped BEV and non-enveloped AAV allowed the specific inactivation of BEV by heat treatment at 60°C for 30 min.28 A simple infection-based method was used to test rAAV activity (Figure 1C). For rAAV2 (293 cell-derived) samples, in 293 cell-based infection assays, both the treated and untreated cells expressed GFP. In Sf9 cell-based infection assays, neither the treated nor untreated cells expressed GFP, indicating that rAAV2 did not infect Sf9 cells. For BEV/Cap2-(ITR-GFP)-Rep supernatant samples, which contained the major secreted BEV and some rAAV, in 293 cell-based infection assays, both the treated and untreated cells expressed GFP to some extent, with the GFP expression of the treated cells being significantly lower than that of the untreated cells. This was because inactive BEV did not express GFP and only some rAAV could express GFP. In Sf9 cell-based infection assays, the untreated cells expressed GFP but the treated cells did not. For BEV/Cap2-(ITR-GFP)-Rep infected Sf9 cell lysate supernatant samples, which contained some non-secreted BEVs and the major rAAV, in 293 cell-based infection assays, both the treated and untreated cells expressed GFP, but GFP expression of the treated cells was slightly lower. This indicates that there was a large amount of rAAVs expressing GFP. In Sf9 cell-based infection assays, the untreated cells expressed GFP, while the treated cells did not express GFP (Figure 1C). To our surprise, the GFP expression level seemed very high under CMV promoter in the Sf9 cell. One reason may be the accumulation of GFP expression along with robust replication of the BEV. Another reason may be that unknown enhancer elements exist upstream of the CMV promoter in the BEV backbone. The results demonstrate that rAAV2 was successfully generated in the novel BEV-infected Sf9 cells.
The Productivity of the New Bac System
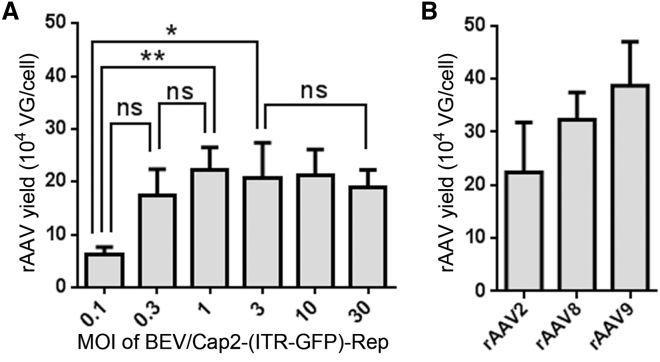
To determine the proper BEV infection ratio for rAAV production, the Sf9 cells cultured in a 6-well plate at 80% confluence were infected with BEV/Cap2-(ITR-GFP)-Rep at an MOI range from 0.1 to 30 with 3-fold dilutions. After 72 hr post-infection, the Sf9 cells were harvested, and the crude cell lysates were used to quantify the DNase-resistant rAAV particles. The yields of rAAV increased from 6.17 ± 1.15 × 104 VG/cell (MOI = 0.1) to 2.23 ± 0.41 × 105 VG/cell (MOI = 1), and they stably maintained a high level at MOIs from 1 to 30 (Figure 2A). The results indicate that the new Bac system exhibits high rAAV yield over a wide range of MOIs. As such, we chose an MOI of 3 for economical and practical considerations.
Figure 2.
Production Properties Analysis of the New Bac System
(A) Analysis of the proper MOI range of BEV/Cap2-(ITR-GFP)-Rep infection. (B) Analysis of the yields for rAAV2, rAAV8, and rAAV9 derived from the Sf9 cells. The yields of rAAVs are determined as vector genomes per cell (VG/cell). All experiments were done in triplicate. Mean ± SD values are presented. Asterisks depict Tukey’s multiple comparison test significance between groups following ANOVA, *p < 0.05 and **p < 0.01; ns, no significant differences.
Further, we tested the yields of rAAV2, rAAV8, and rAAV9 derived from the new Bac system. The suspension-cultured Sf9 cells were infected with the BEVs at an MOI of 3 (about 30 mL, 3 × 106 cells/mL). After 72 hr post-infection, the infected Sf9 cells were harvested and subject to iodixanol gradient purification. For rAAV2, rAAV8, and rAAV9, the yields were 2.24 ± 0.94 × 105 VG/cell, 3.23 ± 0.51 × 105 VG/cell, and 3.87 ± 0.83 × 105 VG/cell, respectively (Figure 2B). Taken together, the results demonstrate that the new Bac system is suitable for large-scale rAAV production.
Stability of BEV/Cap2-(ITR-GFP)-Rep upon Serial Passage
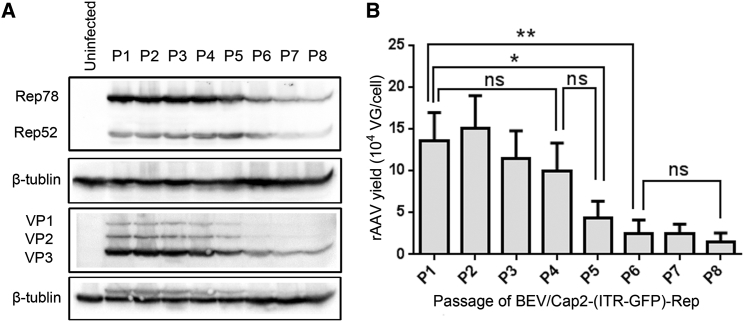
The new BEV/Cap2-(ITR-GFP)-Rep integrated all packaging elements for rAAV production, and so BEV stability is paramount for the scale-up production of rAAV.12 To this end, we amplified the BEV/Cap2-(ITR-GFP)-Rep for serial amplification passages from passage 1 to passage 8. For each passage, BEVs were allowed to infect naive Sf9 cells at an MOI of 0.1, and the culture supernatants were harvested as BEV stocks after 72 hr post-infection. Then, the Sf9 cells cultured in a 6-well plate at 80% confluence were infected with BEV/Cap2-(ITR-GFP)-Rep (MOI = 3) from passage 1 to passage 8. After 72 hr post-infection, the infected Sf9 cells were harvested. To determine whether the Rep and Cap coding sequences were stable in the baculoviruses, the passage samples were analyzed by western blot for Rep and Cap protein expression (Figure 3A). The expression of Rep78/Rep52 and VP1/VP2/VP3 proteins was clearly observed. Stable expression of the Rep and Cap proteins was observed to passage 4. From passage 5 to passage 8, both Rep and Cap expression decreased with passage number.
Figure 3.
Analysis of the BEV Stability upon Serial Passages
(A) Western blot analysis of Rep and Cap protein expression. Passage number is indicated above each lane. The AAV Rep and Cap proteins and endogenous cellular protein β-tubulin are indicated in the left margin. (B) Analysis of the yields of rAAV2 in Sf9 cells upon infection with serial passages of BEV/Cap2-(ITR-GFP)-Rep stocks from passage (P)1 to P8. The yields of rAAVs were determined as vector genomes per cell (VG/cell). All experiments were done in triplicate. Mean ± SD values are presented. Asterisks depict Tukey’s multiple comparison test significance between groups following ANOVA, *p < 0.05 and **p < 0.01; ns, no significant differences.
Because the yield level of rAAV could better reflect the BEV stability, we analyzed the yield level of rAAV upon consecutive passages of the BEV. The data show that the yields of rAAV maintained relatively high levels and had no apparent decrease from passage 1 to passage 4 (all exceeding 105 VG/cell). Beyond passage 4, the yields gradually decreased about 7-fold from passage 4 (1.03 ± 0.30 × 105 VG/cell) to passage 8 (1.47 ± 1.09 × 104 VG/cell) (Figure 3B). The yield level of rAAV correlated with the reduction of the expression level of Rep and Cap proteins. These results demonstrate that the BEV is stable and maintains at least four serial passages without apparent decrease in rAAV yield. The demonstrated BEV stability level could, therefore, support sufficient baculovirus stock amplification for large-scale rAAV production.
Characterization of rAAVs Derived from Novel BEV-Infected Sf9 Cells
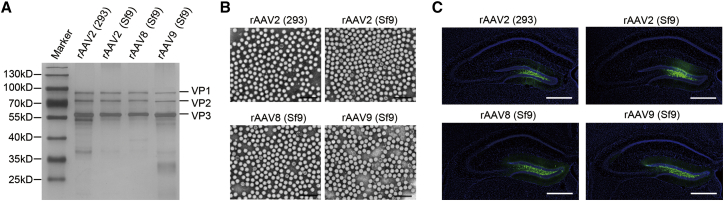
To characterize the rAAVs produced by the new Bac system, iodixanol gradient-purified rAAV2, rAAV8, and rAAV9 were analyzed for their biophysical properties and transduction activity. The rAAV samples were denatured in loading buffer at 95°C for 5 min and then separated by 10% SDS-PAGE. After electrophoresis, gels were silver stained. The data show that the capsid protein compositions of rAAV2, rAAV8, and rAAV9 (Sf9 cell derived) were similar to rAAV2 (293 cell derived). The rAAVs displayed very high purity and presented three bands corresponding to the rAAV capsid proteins VP1 (87 kDa), VP2 (73 kDa), and VP3 (62 kDa), with a ratio of ∼1:1:10 (Figure 4A). Transmission electron microscopy data show that the virions of rAAV2, rAAV8, and rAAV9 (Sf9 cell derived) showed similar structures as rAAV2 (293 cell derived), as relatively uniform spherical particles with a diameter of 20–25 nm. As opposed to full particles, empty particles were distinguished on the basis of the electron-dense central region of the capsid. The data show that full particle contents of the rAAV2, rAAV8, and rAAV9 derived from the new Bac system were about 95.5%, 96.2%, and 94.9% in our preparation (Figure 4B).
Figure 4.
Characterization of Purified rAAVs Derived from BEV/Cap-(ITR-GFP)-Rep-Infected Sf9 Cells
(A) Silver stain of purified rAAVs. HEK293 cell-derived AAV2 (293) and Sf9 cell-derived AAV2 (Sf9), AAV8 (Sf9), and AAV9 (Sf9) were separated by 10% SDS-PAGE, and then they were analyzed by silver staining. Approximately 1 × 1010 VG rAAVs were loaded per lane. (B) Negative staining transmission electron microscopy analysis of purified rAAVs. Representative fields are shown. Original magnification ×100,000. Scale bars, 100 nm. (C) Transduction activity analysis of the rAAVs. Fluorescence microscope images of the mouse brain DG taken from the rAAV-injected mice. Representative fields are shown. Blue, DAPI; green, GFP. Scale bars, 500 μm.
Further, the transduction activities of the rAAVs were analyzed in the brains of C57 mice. The rAAV genome used carried a GFP reporter gene expression cassette driven by the CMV promoter. Similar to the rAAV2- (293 cell derived) injected mice, there was high GFP expression in the dentate gyrus (DG) near the injection sites in the rAAV2-, rAAV8-, and rAAV9- (Sf9 cell derived) injected mice. The fine structure of the GFP-labeled neuron could be seen clearly (Figure 4C). The results demonstrate that the rAAVs derived from the new Bac system exhibited high-quality and high-transduction activity in vivo.
Generation of rAAVs from Novel BEV-Infected Insect Larvae
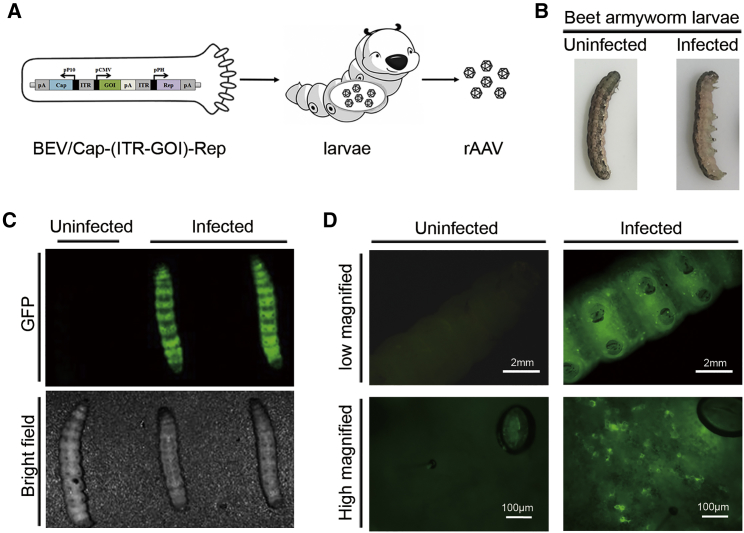
In addition to insect cells, insect larvae have been widely used as a cost-efficient bioreactor for recombinant protein expression.29, 30 The Bac-to-Bac system used in this study was derived from AcMNPV strain E2.27 AcMNPV can infect many kinds of lepidopteran insects, including the beet armyworm (Spodoptera exigua).31, 32 Therefore, we sought to test whether rAAV could be generated from beet armyworm larvae upon infection of the novel BEV/Cap-(ITR-GOI)-Rep (Figure 5A). To test this, fifth-instar beet armyworm larvae were intrahemocelically infected with BEV/Cap2-(ITR-GFP)-Rep. After 3∼4 days post-infection, the infected larvae showed apparent pathological symptoms, which were not observed in uninfected larvae (Figure 5B). We also observed remarkable GFP expression throughout the entire body of the infected larvae, but not in the uninfected larvae (Figure 5C). The GFP-expressing cells could be seen clearly in the subcutaneous tissue of the infected larvae, and they showed uneven distribution along the anteroposterior axis (Figure 5D). The results indicate that the larvae were efficiently infected with the novel BEV.
Figure 5.
Observation of the Beet Armyworm Larvae after Infection by the New BEV/Cap2-(ITR-GFP)-Rep
(A) Schematic of the rAAV produced in beet armyworm larvae upon infection by the new BEV. The functional elements are represented by open boxes and separated by vertical lines. Promoters are represented by arrows. (B) Morphology of uninfected and infected beet armyworm larvae. (C) Fluorescence images of whole-larvae bodies. Upper images are green fluorescence fields, lower images are bright fields. (D) Fluorescence microscopy images of the new BEV/Cap2-(ITR-GFP)-Rep-infected larvae. Low-magnification (1×) scale bars, 2 mm; high-magnification (200×) scale bars, 100 μm. Representative fields are shown.
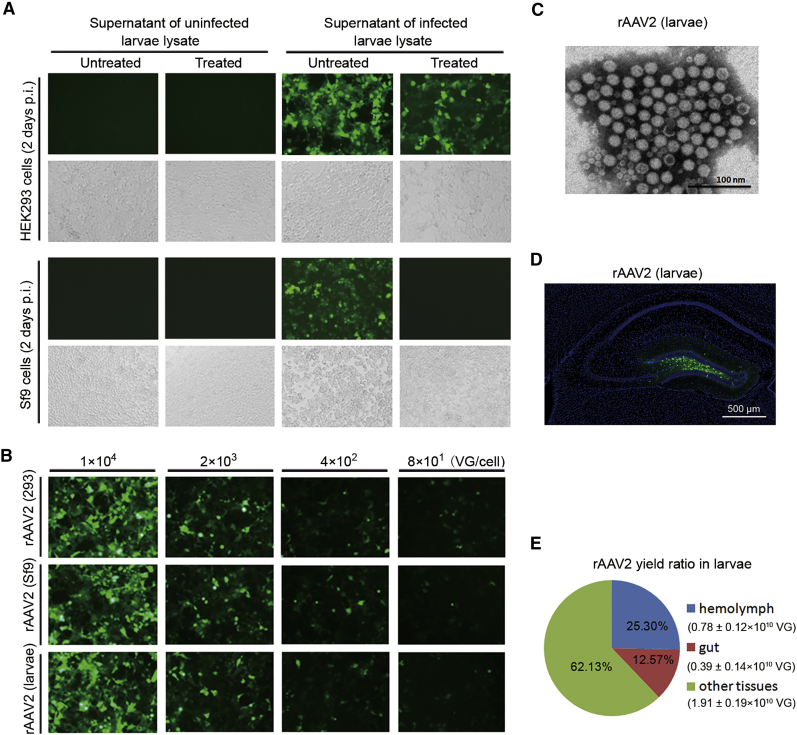
To confirm the production of rAAV2 in the BEV-infected larvae, we used cell-based infection assays to test the rAAV activity as previously described in Figure 1C. For the crude uninfected larvae lysate supernatant samples, in both 293 and Sf9 cell-based infection assays there was no GFP expression (Figure 6A). For the crude infected larvae lysate supernatant samples, in 293 cell-based infection assays, both the treated and untreated cells expressed GFP, but GFP expression of the treated cells was slightly lower. It indicates that there were large amounts of rAAV expressing GFP. In Sf9 cell-based infection assays, untreated cells expressed GFP, but the treated did not (Figure 6A). The results indicate that rAAV2 was successfully produced in the BEV-infected larvae.
Figure 6.
Analysis of rAAV Produced in Beet Armyworm Larvae upon the New BEV/Cap2-(ITR-GFP)-Rep Infection
(A) Fluorescence microscopy observation of HEK293 and Sf9 cells infected with the supernatant of infected or uninfected larvae lysate samples. Treated, heat treatment with 60°C for 30 min; p.i., post-infection. Representative fields are shown; upper images are green fluorescence fields, lower images are bright fields. (B) Comparison of transduction efficiency among rAAV2 derived from HEK293 cells, Sf9 cells, and larvae. HEK293 cells were infected with gradient diluents of rAAVs and examined under a fluorescence microscope 2 days post-infection. (C) Transmission electron microscopy analysis of negatively stained purified rAAV2. Original magnification ×100,000. Scale bars, 100 nm. (D) Transduction activity analysis of larvae-derived rAAV2 in mouse brain DG. Scale bar, 500 μm. Representative fields are shown. (E) Analysis of the yields of rAAV2 in different parts of the larvae. All experiments were done in triplicate and displayed as mean ± SD.
To test the infectivity of the rAAV derived from larvae, we compared the transduction activity of rAAV2 derived from HEK293 cells, Sf9 cells, and larvae in cultured HEK293 cells. The gradient diluent rAAV2 was infected with HEK293 cells, and GFP fluorescence was detected 2 days post-infection. The result indicates that this rAAV2 exhibited similar transduction activities. The purified rAAV2 derived from larvae was then analyzed by transmission electron microscopy. The data show that rAAV2 virions were present as relatively uniform spherical particles with a diameter of 20–25 nm (Figure 6C). This result from larvae-derived rAAV2 is consistent with the transmission electron microscopy (TEM) result from HEK293 cell-derived rAAV2 (Figure 4B). The purified rAAV2 (∼2.0 × 108 VG) derived from larvae was then injected into the mouse brain DG. The data show that there was high GFP expression in the DG and the fine structure of GFP-labeled neurons could be seen clearly (Figure 6D). These results demonstrate that the rAAVs derived from larvae have high-transduction activity in vivo.
Next, we assessed the rAAV2 yield in the novel BEV-infected beet armyworm larvae. The data showed that the average rAAV2 yield per larvae was 2.75 ± 1.66 × 1010 VG in our preparation. Further, we analyzed the rAAV2 yield in different parts of the larvae. The data showed that there were about 0.78 ± 0.12 × 1010 VG, 0.39 ± 0.14 × 1010 VG, and 1.91 ± 0.19 × 1010 VG in the hemolymph (25.30%), gut (12.57%), and other tissues (62.13%), respectively (Figure 6E). These results suggest that the larvae are a cost-efficient platform for rAAV production.
Discussion
In 2002, Urabe et al. demonstrated that the rAAV vector could reproduce in cultured Sf9 cells upon triple BEV infection. The results suggest that insect cells support the replication of AAV, and baculovirus infection could supply the helper function, although the detailed mechanism is not known.18 Since then, further improvements have been made in two key aspects: (1) to improve genetic stability of the BEVs, and (2) to reduce the complexity of the Bac system.19, 22 In short, AAV Cap and Rep helper genes were incorporated into a single BEV or integrated into Sf9 packaging cell lines, and the two-Bac systems and Sf9 packaging cell line-based OneBac systems were generated.20, 21, 22 However, current Bac systems still remain complicated and cost-intensive, and they lack versatility and flexibility.
The ideal solution is developing an efficient BEV integrating all the necessary packaging elements for rAAV production. As such, it is a difficult challenge to construct a BEV to such specifications. One difficulty is how to organize the Cap and Rep genes as well as the rAAV genome (ITR-GOI) in a single BEV while ensuring that the organization does not disrupt one another’s functions. Another main concern is the genetic stability of the highly integrated BEV.33 Moreover, the AAV ITR sequences are susceptible to AAV Rep-mediated cleavage.34 Whether the progeny BEV remain integrated in the situation of the simultaneous existence of the expression of the Rep gene and rAAV packaging is also of concern. Thus, the designs of the BEV constructs should consider two rules. First, the Rep and Cap genes and (ITR-GOI) must be optimally organized without disrupting one another’s functions. Second, the constructs should be versatile and flexible for different kinds of rAAV production. To this end, we integrated them into a single pFD shuttle vector, thus facilitating BEV construction through Bac-to-Bac operation. Based on previous achievements of the Bac systems, we employed the ribosome leaky-scanning mechanism to express AAV Rep and Cap proteins downstream the PH and P10 promoters in the pFD vector, respectively.21 In the pFD vector, the PH and P10 promoters are head-to-head linked by a short DNA fragment, which contains an AccIII cut site. This site is suitable for the insertion of the (ITR-GOI). To minimize the adverse impact of insertion, we brought in short linker DNA fragments (about 80∼140 bp) outside of each ITR through PCR amplification. Finally, the functional BEV/Cap-(ITR-GFP)-Rep was produced from the Bacmid-transfected Sf9 cells (Figures 1A and 1B). The rAAVs were successfully generated from the Sf9 cells upon BEV/Cap-(ITR-GFP)-Rep infection (Figure 1C).
The new Bac system displayed a wide range of MOIs for high-level rAAV production (Figure 3A). This property makes it suitable for optimizing the fermentation process at high cell density.17 The yield levels of rAAV2, rAAV8, and rAAV9 derived from the new Bac systems exceeded 105 VG/cell (Figure 3B), which is close to the current best states of large-scale rAAV production systems.22, 23 To our delight, the BEV/Cap-(ITR-GFP)-Rep was shown to be stable over at least four amplification passages. Moreover, the expression of Rep and Cap proteins maintained stable levels during passages 1–4 and gradually decreased from passages 5–8, which is correlated with the yield levels of rAAV (Figure 4). Compared with the previously reported BEV/Cap-Rep, which showed stable expression of the AAV Rep and Cap proteins over seven passages,21 the apparently lower stability of BEV/Cap-(ITR-GFP)-Rep could due to the additional ITR-containing component, as there was a notable loss of the ITR-transgene cassette-containing baculovirus over the five passages, as previously reported.19 Further improvements to the stability of the new BEV are needed, and a recent study on lef5 should be noted in this effort.35 It is noteworthy that the new Bac system produced rAA2, rAAV8, and rAAV9 viral particles with high integrity ratios (exceeding 90% full particles) in our preparation (Figure 4B). In contrast, for the three-Bac and the two-Bac systems, the low integrity ratios of the rAAVs are likely due to improper co-infection ratios of the BEVs.36 The result suggests that single BEV/Cap-(ITR-GOI)-Rep infection has a unique advantage for high-quality rAAV production.
In addition to cultured insect cells, the insect larvae have been widely used for exogenous protein expression.29 Although rAAV has been successfully produced in cultured Sf9 cells for years, there are as of yet no reports that rAAV has been produced in insect larvae. This is because, unlike in cultured Sf9 cells, it is hard to control multi-BEV co-infection with insect larvae. Also, unlike in the stable Sf9 packaging cell line, it is very difficult to gain transgenic insects suitable for the production of rAAVs. The successful production of functional BEV/Cap-(ITR-GOI)-Rep, therefore, paves the way for efficient production of rAAV in insect larvae. A further consideration is that the beet armyworm is the natural host of the AcMNPV-derived BEV/Cap-(ITR-GOI)-Rep in this study. In the present study, we tested whether rAAV could be produced in the BEV/Cap-(ITR-GOI)-Rep-infected beet armyworm larvae. We found that the BEV replicated efficiently throughout the entire larval body after 3∼4 days of intrahemocelical infection (Figure 5). The rAAV2 was successfully generated in BEV-infected beet armyworm larvae (Figure 6A). The larvae-derived rAAV2 show similar in vitro and in vivo transduction activity to the HEK293- and Sf9-derived rAAVs (Figures 6B and 6D). The average yield of rAAV2 per larvae was observed to be 2.75 ± 1.66 × 1010 VG in our preparation. Unlike secreted proteins, which mainly exist in the larvae hemolymph, about 74.7% of the rAAV existed in the larvae tissues with about only 25.3% of the rAAV existing in the larvae hemolymph (Figure 6E). An efficient purification method of rAAVs produced in insect larvae still needs to be further developed.29 Furthermore, the BEV used in this study is derived from polyhedron-negative AcMNPV (polh−) and lacked oral infectivity.26 Compared to intrahemocoelical injection, oral infection is more suitable for scale-up production of rAAV in insect larvae due to less labor-intensive manual operation.37 Thus, oral infection might be better for large-scale production of rAAV in insect larvae. Moreover, to find the ideal insect larvae bioreactor for the production of rAAV, it will be worth testing the rAAV productivity with different types of recombinant baculovirus and insect larvae in the future, such as recombinant Bombyx mori nucleopolyhedrovirus (BmNPV) and its host silkworm (Bombyx mori) larvae, which have been widely exploited.38
In conclusion, high-quality rAAVs were produced in the new BEV/Cap-(ITR-GFP)-Rep-infected Sf9 cells. The yields of rAAV2, rAAV8, and rAAV9 exceeded 105 VG/cell, which is close to the current best states of large-scale rAAV production systems. The new BEV is stable for at least four amplification passages. The new Bac system is ideally suitable for large-scale rAAV production. In addition, it is the first demonstration of rAAV2 production in beet armyworm larvae using the new BEV infection. The rAAV2 derived from larvae show a structure similar to the HEK293 cell-derived rAAV2 and high-transduction activity, both in cultured HEK293 cells and mouse brain DG. The average rAAV2 yield exceeded 1010 VG per larvae, and the rAAV2 was seen to mainly exist in larval tissues. Thus, this study exploits a potential cost-efficient platform for rAAV production in insect larvae.
Materials and Methods
Plasmid Construction
An artificially synthesized AAV2 Rep gene expressing a bifunctional Rep78- and Rep52-encoding mRNA was amplified with primers Rep-Bgl-F and Rep-Xba-R, cloned into pFD plasmid between BamH1 and Xba1 cut sites to create the pFD/Rep.21 The Cap2, Cap8, and Cap9 genes were amplified from pAAV-RC2, pAAV-RC8, and pAAV-RC9 with primers Cap2-Xma-F and Cap2-Nhe-R, Cap8-Xma-F and Cap8-Nhe-R, and Cap9-Xma-F and Cap9-Nhe-R, and then they were cloned into pFD between Xma1 and Nhe1 cut sites to create the pFD/Cap2, pFD/Cap8, and pFD/Cap9, respectively.18, 21 The ITR-(L) and ITR-(R) fragments were amplified from pAAV-MCS with primers ITR-KpnM-F and Sac1-R or primers Kpn-Sac1-F and ITR-SpeM-R, and then they were ligated into psimpleT-ITR. A GFP reporter gene was amplified by primers EcoR-GFP-F and GFP-BamH-R, and then it was inserted into psimpleT-ITR between EcoR1 and BamH1 cut sites to create psimpleT-(ITR-GFP). The (ITR-GFP) fragment was inserted into pFD/Cap2 between Mro1 and Spe1 cut sites to create pFD/Cap2-(ITR-GFP). The PH-Rep fragment was amplified from pFD/Rep with primers Rep-Spe-F and Rep-Xba-R, and it was inserted into pFD/Cap2-(ITR-GFP) at the Spe1 cut site to create pFD/Cap2-(ITR-GFP)-Rep. The Cap2 gene in pFD/Cap2-(ITR-GFP)-Rep was replaced by Cap8 or Cap9 genes from pFD/Cap8 or pFD/Cap9 through Pac1 and Nhe1 cut sites to obtain pFD/Cap8-(ITR-GFP)-Rep or pFD/Cap9-(ITR-GFP)-Rep, respectively. Recombinant baculoviruses were generated following the Bac-to-Bac system protocol (Invitrogen). All constructs were confirmed by sequencing. Primers used for PCR are shown in Table S1.
Cell and Insect Larvae Culture
HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Adherent culture of Sf9 cells was maintained in plate cultures containing Grace’s Insect Medium (Gibco) supplemented with 10% FBS, at 28°C. Suspension culture of Sf9 cells was maintained in shake flask cultures containing Sf-900II SFM (Gibco), at 28°C. Media were supplemented with 100 μg/mL streptomycin and 100 U/mL penicillin. The beet armyworm (Spodoptera exigua) larvae were raised at 25°C under 14-hr:10-hr light:dark cycle with an artificial diet.
Larvae Sample Preparation and Observation
For beet armyworm larvae injection, the fifth-instar larvae were infected by intrahemocelical injection with 10 μL (∼1.0 × 106 VG) BEV solutions (or sterile PBS buffer as uninfected control) near the proleg (forward from the body cavity), as previously described.39 After 3∼4 days post-infection, infected larvae were collected and washed three times with PBS buffer (pH 7.4). For larval tissue isolation, the larvae were narcotized on cold ice, and then they were surgically dissected to collect the hemolymph, gut, and other tissues (mainly including the fat body and epidermis). The larvae or tissue samples were then homogenized in PBS buffer, and then they were subjected to three freeze-thaw cycles with liquid nitrogen and a 37°C water bath, followed by centrifugation at 5,000 × g for 10 min. After centrifugation, the larvae or the tissue crude lysate supernatants were immediately subject to downstream operation or stored at −80°C until use. The rest of the extraction procedure for the larvae-derived rAAV was similar to that used for Sf9 cell-derived rAAV. For the observation of larvae, the larvae were narcotized on cold ice, and whole-larvae fluorescence imaging was carried out by an in vivo imaging system (CRi Maestro2). Low-magnification imaging was carried out by stereo microscope (Olympus SZX16), and high-magnification imaging was carried out by inverted microscope (Olympus IX71).
rAAV Production and Purification
rAAV production in HEK293 cells was performed as previously described.40 Briefly, HEK293 cells at 80% confluence were co-transfected by polyethylenimine (PEI) method with plasmids pAAV-RC2, pAAV-helper, and pAAV-GFP. Cells were harvested 3 days post-transfection. For Sf9 cells, adherent cultured insect cells at 80% confluence were infected with BEV at an MOI of 3. Suspension-cultured insect cells at 3 × 106 cells/mL density were infected with BEV (MOI = 3). Cells were harvested 3 days post-infection. HEK293 cells or Sf9 cells were suspended in lysis buffer (50 mM Tris and 2 mM MgCl2 [pH 7.5]) at a cell density of 2 × 107 cells/mL. Cells were lysed by three freeze-thaw cycles using liquid nitrogen and a 37°C water bath. The crude cells lysates were mixed with 50 U/mL benzonase (Sigma) with the addition of 150 mM NaCl for 1 hr at 37°C. Then, the mixture was centrifuged at 2,500 × g for 15 min and filtered. The supernatant fractions were used for downstream purification.
An iodixanol step density gradient was prepared as described previously.41, 42 Briefly, 15%, 25%, 40%, and 58% iodixanol were diluted in PBS-MK Buffer (1× PBS, 1 mM MgCl2, and 2.5 mM KCl) from 60% iodixanol (OptiPrep; Sigma). The cell lysate samples and iodixanol solution were successively underlaid in 39 mL Quick-Seal tubes (Beckman Coulter). After centrifugation in a 70 Ti rotor for 2 hr at 63,000 rpm and 18°C, the fractions obtained from the 40% phase were dialyzed against PBS and then ultrafiltered and concentrated, using Amicon Ultra-15 centrifugal filter units (MWCO, 100 kDa; Merck Millipore). The purified rAAVs were aliquoted and stored at −80°C.
Virus Titration
Virus titers were determined by qPCR assay with the iQ SYBR Green Supermix kit (Bio-Rad). A standard curve was obtained using 10-fold serial dilutions of plasmids. The BEV samples were treated with a modified alkaline polyethylene glycol (PEG)-based method as previously described.22 The preparations with highly purified rAAV vector or rAAV in crude lysate supernatants were performed as previously described.23 For the titration of the rAAV derived from crude lysates of Sf9 cell, the crude lysates were heated to inactivate the BEV before benzonase treatment. The quantitative primers for BEV and rAAV were Bac-F and Bac-R and CMV-F and CMV-R, respectively. Primers used for qPCR are shown in Table S1.
Western Blot and Silver Staining
Western blot analysis was performed as previously described.21 Samples were separated by using 10% SDS-PAGE. Rep proteins were detected using an anti-Rep monoclonal antibody (clone 303.9, Fitzgerald) at a 1:100 dilution. Cap proteins were detected using an anti-AAV VP1/VP2/VP3 monoclonal antibody (clone B1, Progen) at a 1:100 dilution. A monoclonal antibody against β-tubulin (Proteintech) was used at a 1:1,000 dilution. Secondary detection antibodies were used at a 1:2,000 dilution. For silver staining, fixed gels were stained using the Fast Silver Stain Kit (Beyotime) according to the manufacturer’s protocol.
Transmission Electron Microscopy
The virus particles were assessed by negative stain and transmission electron microscopy. An approximately 10-μL sample was placed on a 400-mesh carbon-coated copper grid and incubated for 2 min, quickly removed, and air dried. The grid was then stained with 2% phosphotungstic acid (pH 7.4) for 2 min, washed with a drop of water, and then air dried again. Experiments were performed with a transmission electron microscope (Hitachi H-7000A).
rAAV In Vivo Property Assay
All animal experiment protocols were approved by Animal Care and Use Committees at the Wuhan Institute of Physics and Mathematics, the Chinese Academy of Sciences. 8-week-old male C57BL/6 mice were used for the virus transduction assay. Briefly, the mice were anesthetized with chloral hydrate and secured in a stereotaxic apparatus. The skull of the targeted site was thinned by a dental drill. A syringe (10 μL) connected to a glass micropipette with a 10- to 15-μm diameter tip was used for virus injection. The rAAV (∼2.0 × 108 VG) in a volume of 0.2 μL was stereotaxically microinjected into the DG, using the coordinates 1.70 mm posterior to bregma, 1.00 mm lateral from midline, and 2.0 mm dorsoventral relative to bregma. The syringe was left in place for 10 min following injection to minimize diffusion. At 3 weeks after injection, the mice were sacrificed. Mice were deeply anesthetized with an overdose of chloral hydrate and transcardially perfused with 0.9% saline solution followed by 4% paraformaldehyde (PFA) solution. The brains were removed, then fixed in 4% PFA overnight, and dehydrated in 30% sucrose before being sectioned into 40-μm slices (Thermo Fisher Scientific). Fixed slices were stained with DAPI and imaged by virtual slide microscope (Olympus VS120).
Author Contributions
Y.W. conceived and performed experiments, analyzed the data, and wrote and edited the manuscript. L.J. performed experiments and analyzed the data. H.G. and Z.H. carried out viral vector production. T.Y. carried out animal studies. X.H. contributed to the conception and design of experiments. K.L. contributed to larvae experiments. F.X. designed experiments, supervised the project, and wrote and edited the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We are grateful to Liting Luo and Lingling Xu (Core Facility Center, Wuhan Institute of Physics and Mathematics) for technical assistance in centrifugation and microscope imaging, and Ding Gao (Core Facility Center, Wuhan Institute of Virology) for technical assistance in electronic microscopy. This work was supported financially by the National Natural Science Foundation of China (31500868 to Y.W.), the National Basic Research Program (973 Program) of China (2015CB755600 to F.X.), Strategic Priority Research Program (B) (XDB0205005 to F.X.), and the National Natural Science Foundation of China (31771156 to F.X.).
Footnotes
Supplemental Information includes one table and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.05.005.
Contributor Information
Yang Wu, Email: yangwu@wipm.ac.cn.
Fuqiang Xu, Email: fuqiang.xu@wipm.ac.cn.
Supplemental Information
References
- 1.Betley J.N., Sternson S.M. Adeno-associated viral vectors for mapping, monitoring, and manipulating neural circuits. Hum. Gene Ther. 2011;22:669–677. doi: 10.1089/hum.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller C., Flotte T.R. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- 3.Mingozzi F., High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 4.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varenika V., Kells A.P., Valles F., Hadaczek P., Forsayeth J., Bankiewicz K.S. Controlled dissemination of AAV vectors in the primate brain. Prog. Brain Res. 2009;175:163–172. doi: 10.1016/S0079-6123(09)17511-8. [DOI] [PubMed] [Google Scholar]
- 6.Pierce E.A., Bennett J. The Status of RPE65 Gene Therapy Trials: Safety and Efficacy. Cold Spring Harb. Perspect. Med. 2015;5:a017285. doi: 10.1101/cshperspect.a017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacLaren R.E., Groppe M., Barnard A.R., Cottriall C.L., Tolmachova T., Seymour L., Clark K.R., During M.J., Cremers F.P., Black G.C. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant L.M., Christopher D.M., Giles A.R., Hinderer C., Rodriguez J.L., Smith J.B., Traxler E.A., Tycko J., Wojno A.P., Wilson J.M. Lessons learned from the clinical development and market authorization of Glybera. Hum. Gene Ther. Clin. Dev. 2013;24:55–64. doi: 10.1089/humc.2013.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayuso E., Mingozzi F., Bosch F. Production, purification and characterization of adeno-associated vectors. Curr. Gene Ther. 2010;10:423–436. doi: 10.2174/156652310793797685. [DOI] [PubMed] [Google Scholar]
- 11.Samulski R.J., Chang L.S., Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cecchini S., Negrete A., Kotin R.M. Toward exascale production of recombinant adeno-associated virus for gene transfer applications. Gene Ther. 2008;15:823–830. doi: 10.1038/gt.2008.61. [DOI] [PubMed] [Google Scholar]
- 13.Grieger J.C., Soltys S.M., Samulski R.J. Production of Recombinant Adeno-associated Virus Vectors Using Suspension HEK293 Cells and Continuous Harvest of Vector From the Culture Media for GMP FIX and FLT1 Clinical Vector. Mol. Ther. 2016;24:287–297. doi: 10.1038/mt.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H., Xie J., Xie Q., Wilson J.M., Gao G. Adenovirus-adeno-associated virus hybrid for large-scale recombinant adeno-associated virus production. Hum. Gene Ther. 2009;20:922–929. doi: 10.1089/hum.2009.125. [DOI] [PubMed] [Google Scholar]
- 15.Clément N., Knop D.R., Byrne B.J. Large-scale adeno-associated viral vector production using a herpesvirus-based system enables manufacturing for clinical studies. Hum. Gene Ther. 2009;20:796–806. doi: 10.1089/hum.2009.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamson-Small L., Potter M., Falk D.J., Cleaver B., Byrne B.J., Clément N. A scalable method for the production of high-titer and high-quality adeno-associated type 9 vectors using the HSV platform. Mol. Ther. Methods Clin. Dev. 2016;3:16031. doi: 10.1038/mtm.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galibert L., Merten O.W. Latest developments in the large-scale production of adeno-associated virus vectors in insect cells toward the treatment of neuromuscular diseases. J. Invertebr. Pathol. 2011;107(Suppl):S80–S93. doi: 10.1016/j.jip.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Urabe M., Ding C., Kotin R.M. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- 19.Kohlbrenner E., Aslanidi G., Nash K., Shklyaev S., Campbell-Thompson M., Byrne B.J., Snyder R.O., Muzyczka N., Warrington K.H., Jr., Zolotukhin S. Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol. Ther. 2005;12:1217–1225. doi: 10.1016/j.ymthe.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H. Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol. Ther. 2008;16:924–930. doi: 10.1038/mt.2008.35. [DOI] [PubMed] [Google Scholar]
- 21.Smith R.H., Levy J.R., Kotin R.M. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol. Ther. 2009;17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aslanidi G., Lamb K., Zolotukhin S. An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc. Natl. Acad. Sci. USA. 2009;106:5059–5064. doi: 10.1073/pnas.0810614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mietzsch M., Grasse S., Zurawski C., Weger S., Bennett A., Agbandje-McKenna M., Muzyczka N., Zolotukhin S., Heilbronn R. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1-12 vectors for gene therapy. Hum. Gene Ther. 2014;25:212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mietzsch M., Casteleyn V., Weger S., Zolotukhin S., Heilbronn R. OneBac 2.0: Sf9 cell lines for production of AAV5 vectors with enhanced infectivity and minimal encapsidation of foreign DNA. Hum. Gene Ther. 2015;26:688–697. doi: 10.1089/hum.2015.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mietzsch M., Hering H., Hammer E.M., Agbandje-McKenna M., Zolotukhin S., Heilbronn R. OneBac 2.0: Sf9 Cell Lines for Production of AAV1, AAV2, and AAV8 Vectors with Minimal Encapsidation of Foreign DNA. Hum. Gene Ther. Methods. 2017;28:15–22. doi: 10.1089/hgtb.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luckow V.A., Lee S.C., Barry G.F., Olins P.O. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayres M.D., Howard S.C., Kuzio J., Lopez-Ferber M., Possee R.D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y.K., Yang C.J., Liu C.L., Shen C.R., Shiau L.D. Using a fed-batch culture strategy to enhance rAAV production in the baculovirus/insect cell system. J. Biosci. Bioeng. 2010;110:187–193. doi: 10.1016/j.jbiosc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Targovnik A.M., Arregui M.B., Bracco L.F., Urtasun N., Baieli M.F., Segura M.M., Simonella M.A., Fogar M., Wolman F.J., Cascone O., Miranda M.V. Insect Larvae: A New Platform to Produce Commercial Recombinant Proteins. Curr. Pharm. Biotechnol. 2016;17:431–438. doi: 10.2174/138920101705160303163947. [DOI] [PubMed] [Google Scholar]
- 30.Maeda S., Kawai T., Obinata M., Fujiwara H., Horiuchi T., Saeki Y., Sato Y., Furusawa M. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature. 1985;315:592–594. doi: 10.1038/315592a0. [DOI] [PubMed] [Google Scholar]
- 31.Guo T., Wang S., Guo X., Lu C. Productive infection of Autographa californica nucleopolyhedrovirus in silkworm Bombyx mori strain Haoyue due to the absence of a host antiviral factor. Virology. 2005;341:231–237. doi: 10.1016/j.virol.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 32.Wu C.Y., Chen Y.W., Lin C.C., Hsu C.L., Wang C.H., Lo C.F. A new cell line (NTU-SE) from pupal tissues of the beet armyworm, Spodoptera exigua (Lepidoptera: Noctuidae), is highly susceptible to S. exigua multiple nucleopolyhedrovirus (SeMNPV) and Autographa californica MNPV (AcMNPV) J. Invertebr. Pathol. 2012;111:143–151. doi: 10.1016/j.jip.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 33.Pijlman G.P., van Schijndel J.E., Vlak J.M. Spontaneous excision of BAC vector sequences from bacmid-derived baculovirus expression vectors upon passage in insect cells. J. Gen. Virol. 2003;84:2669–2678. doi: 10.1099/vir.0.19438-0. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Brister J.R., Im D.S., Muzyczka N. Characterization of the adenoassociated virus Rep protein complex formed on the viral origin of DNA replication. Virology. 2003;313:364–376. doi: 10.1016/s0042-6822(03)00340-4. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-Solís M., Jakubowska A.K., Herrero S. Expression of the lef5 gene from Spodoptera exigua multiple nucleopolyhedrovirus contributes to the baculovirus stability in cell culture. Appl. Microbiol. Biotechnol. 2017;101:7579–7588. doi: 10.1007/s00253-017-8495-y. [DOI] [PubMed] [Google Scholar]
- 36.Urabe M., Nakakura T., Xin K.Q., Obara Y., Mizukami H., Kume A., Kotin R.M., Ozawa K. Scalable generation of high-titer recombinant adeno-associated virus type 5 in insect cells. J. Virol. 2006;80:1874–1885. doi: 10.1128/JVI.80.4.1874-1885.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guijarro-Pardo E., Gómez-Sebastián S., Escribano J.M. In vivo production of recombinant proteins using occluded recombinant AcMNPV-derived baculovirus vectors. J. Virol. Methods. 2017;250:17–24. doi: 10.1016/j.jviromet.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Kato T., Kajikawa M., Maenaka K., Park E.Y. Silkworm expression system as a platform technology in life science. Appl. Microbiol. Biotechnol. 2010;85:459–470. doi: 10.1007/s00253-009-2267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Millán A.F., Gómez-Sebastián S., Nuñez M.C., Veramendi J., Escribano J.M. Human papillomavirus-like particles vaccine efficiently produced in a non-fermentative system based on insect larva. Protein Expr. Purif. 2010;74:1–8. doi: 10.1016/j.pep.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Grieger J.C., Choi V.W., Samulski R.J. Production and characterization of adeno-associated viral vectors. Nat. Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 41.Strobel B., Miller F.D., Rist W., Lamla T. Comparative Analysis of Cesium Chloride- and Iodixanol-Based Purification of Recombinant Adeno-Associated Viral Vectors for Preclinical Applications. Hum. Gene Ther. Methods. 2015;26:147–157. doi: 10.1089/hgtb.2015.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.