Abstract
Epithelial ovarian cancer (EOC) is the fifth leading cause of cancer mortality in American women. Normal ovarian physiology is intricately connected to small GTP binding proteins of the Ras superfamily (Ras, Rho, Rab, Arf, and Ran) which govern processes such as signal transduction, cell proliferation, cell motility, and vesicle transport. We hypothesized that common germline variation in genes encoding small GTPases is associated with EOC risk. We investigated 322 variants in 88 small GTPase genes in germline DNA of 18,736 EOC patients and 26,138 controls of European ancestry using a custom genotype array and logistic regression fitting log-additive models. Functional annotation was used to identify biofeatures and expression quantitative trait loci that intersect with risk variants. One variant, ARHGEF10L (Rho guanine nucleotide exchange factor 10 like) rs2256787, was associated with increased endometrioid EOC risk (OR = 1.33, p = 4.46 x 10−6). Other variants of interest included another in ARHGEF10L, rs10788679, which was associated with invasive serous EOC risk (OR = 1.07, p = 0.00026) and two variants in AKAP6 (A-kinase anchoring protein 6) which were associated with risk of invasive EOC (rs1955513, OR = 0.90, p = 0.00033; rs927062, OR = 0.94, p = 0.00059). Functional annotation revealed that the two ARHGEF10L variants were located in super-enhancer regions and that AKAP6 rs927062 was associated with expression of GTPase gene ARHGAP5 (Rho GTPase activating protein 5). Inherited variants in ARHGEF10L and AKAP6, with potential transcriptional regulatory function and association with EOC risk, warrant investigation in independent EOC study populations.
Introduction
In 2017, in the United States, more than 21,000 women were expected to be diagnosed with epithelial ovarian cancer (EOC), and more than 14,000 women were predicted to die from the disease.[1] EOC is heterogeneous and therefore classified into major histological subtypes of invasive disease—serous, endometrioid, clear cell, and mucinous–and two histological subtypes of borderline disease–serous and mucinous. These histological subtypes have differences in genetic and epidemiologic risk factors, molecular events during oncogenesis, response to chemotherapy, and prognosis.[2]
Approximately 20% of the familial component of EOC risk is attributable to high-to-intermediate risk gene mutations.[3] In European populations, genome-wide association studies (GWAS) have identified more than 30 EOC susceptibility alleles, as reviewed previously.[4] Known common genetic variants explain 3.9% of the inherited component of EOC risk, and additional susceptibility loci are likely to exist, particularly for the less common, non-serous histological subtypes.
Normal ovarian physiology is intricately connected to tightly regulated small GTP binding proteins of the Ras superfamily (Ras, Rho, Rab, Ral, Arf, and Ran) which regulate key cellular processes such as signal transduction, cell proliferation, cell motility, and vesicle transport.[5] These proteins function in a highly coordinated manner through signaling networks and feedback loops within and among the small GTPase subfamilies.[6] The Rab and Ral GTPases are thought to function in membrane trafficking in exocyst assembly and vesicle-tethering processes;[7, 8] Rho-related proteins function to integrate extracellular signals with specific targets regulating cell morphology, cell aggregation, tissue polarity, cell motility and cytokinesis.[5] Ras family genes cycle between their inactive GDP forms in the cytoplasm and the active GTP-bound forms on the plasma membrane and are associated with signaling pathways contributing to normal and aberrant cell growth.[9]
As regulation of the RAS signal transduction pathway involves a highly complex, highly polymorphic machinery of genes, we conducted a large-scale candidate pathway association study, hypothesizing that variation in small GTPase genes is associated with EOC risk.
Materials and methods
Variant selection
RAS pathway genes were selected based on the Cancer Genome Anatomy Project and review of the published literature (www.pubmed.gov). Within 115 candidate genes, 6103 single nucleotide polymorphism (SNPs) were interrogated in early GWAS analysis of 7931 EOC patients and 9206 controls;[10] 339 SNPs in 88 of these genes showed nominal evidence of association with risk of EOC or of serous EOC (p<0.05 using all participants or North American participants only)[10] and were targeted in the present analysis (S1 Table).
Study participants and genotyping
We studied 18,736 EOC patients (10,316 of serous histology) and 26,138 controls who participated in Ovarian Cancer Association Consortium studies; all participants were of European ancestry.[11] This included participants from the GWAS which was used for variant selection (described above)[10] and an additional 10,243 patients and 16,932 controls. Genotyping used a custom Illumina Infinium array. [11] SNPs were excluded according to the following criteria: no genotype call; monomorphism; call rate less than 95% and minor allele frequency > 0.05 or call rate less than 99% with minor allele frequency < 0.05; evidence of deviation of genotype frequencies from Hardy-Weinberg equilibrium (p < 10−7); greater than 2% discordance in duplicate pairs. Overall, 322 small GTPase gene SNPs were genotyped and passed QC; numbers of participants with data for each SNP vary, as some DNA samples failed QC for particular SNPs. This study was reviewed and approved by the Mayo Clinic Institutional Review Board as protocol 1367–05.
Genetic association
We followed STREGA guidelines for genetic association studies.[12] Unconditional logistic regression treating the number of minor alleles carried as an ordinal variable (log-additive model) was used to evaluate the association between each SNP and EOC risk adjusted for age, study site, and principal components to account for residual differences in European ancestry. Six series of analyses were conducted considering the following groups: all invasive EOC combined, each of the four main invasive histological subtypes (serous, endometrioid, clear cell and mucinous), and all borderline tumors combined. No corrections were made for multiple testing.
Functional annotation
For SNPs of interest, dbSUPER [13] and Haploreg v4.1[14] were used to evaluate publicly available data for variant overlap with human super-enhancers,[15] known expression quantitative trait loci (eQTL), GWAS hits, and other regulatory marks. In addition, we assessed correlations between germline genotype with tumor expression levels (eQTL analysis) using 312 Mayo Clinic patients (226 serous, 54 endometrioid, 22 clear cell, 5 mucinous, and 5 of other histological subtypes). Expression data were obtained using fresh frozen tumor RNA and Agilent whole human genome 4×44 expression arrays and were analyzed in the form of log ratios of signals from individual tumors compared to signals from a reference mix of 106 tumor samples[16, 17] versus signals from a reference mix of 106 tumor samples[16, 17]. Expression levels for minor allele carriers versus non-carriers were compared using the Wilcoxon rank sum statistic.
Results and discussion
Demographic and clinical characteristics of the study sample (18,736 EOC patients and 26,138 controls) have been described previously.[11] In brief, compared to controls, patients were older, attained menarche at older ages, and had higher body mass index. As expected, most tumors (57.6%) were of serous histology with 14.2% endometrioid, 7.1% clear cell, 6.5% mucinous, and 14.6% other/unknown.
From among 322 SNPs in 88 RAS pathway small GTPase genes, we observed that 99 SNPs in 43 genes were nominally associated with EOC risk (p<0.05) (S2 Table). These associations were from six separate analyses that evaluated all patients with invasive disease, patients with one of the four main invasive histological subtypes, serous [n = 8,372], endometrioid [n = 2,068], clear cell [n = 1,025] and mucinous [n = 943], as well as patients with borderline tumors.
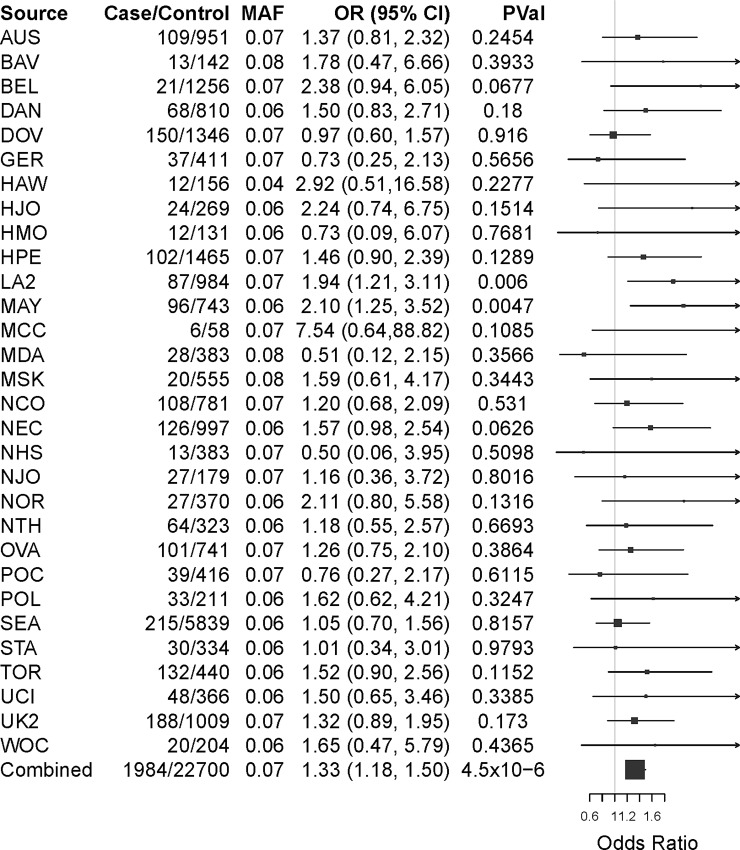
In ARHGEF10L, which encodes the Rho guanine nucleotide exchange factor 10-like protein, SNP rs2256787 was associated with invasive endometrioid EOC risk (OR = 1.33, 95% CI: 1.18–1.50, p = 4.5x10-6) (Table 1). (Fig 1) shows the ORs and 95% CIs associated with the G allele at this SNP overall and by contributing study.
Table 1. Association of variants in small GTPase genes with epithelial ovarian cancer risk (p-value<10−4) and functional annotation.
| Genetic Association | Functional Annotation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Chr:Position | Alleles |
MAF | Histology | OR (95% CI) | P-value | Conserved site | eQTL | Tissues with enhancer histone mark | Tissues with DNAse site | In super-enhancer |
| ARHGEF10L | rs2256787 | 1:17,765,403 | A/C | 0.07 | Endometrioid | 1.33 (1.18–1.50) | 4.5 x 10−6 | No | No | ESC, ESDR, IPSC, FAT, STRM, BRST, BRN, SKIN, VAS, LIV, GI, HRT, MUS, LNG, OVRY, PANC | None | Yes |
| rs10788679 | 1:17,789,549 | A/G | 0.42 | Serous | 1.07 (1.03–1.11) | 2.6 x 10−4 | No | No | None | None | Yes | |
| AKAP6 | rs1955513 | 14:32,245,693 | C/A | 0.07 | All invasive | 0.90 (0.85–0.95) | 3.3 x 10−4 | Yes | No | FAT, SKIN, VAS, BRN, MUS, GI, BLD | SKIN,MUS,MUS,THYM,BLD | No |
| rs927062 | 14:32,164,800 | G/A | 0.21 | All invasive | 0.94 (0.90–0.97) | 5.9 x 10−4 | No | Yes, ARHGAP5 | None | GI | No | |
SNP, single nucleotide polymorphism; alleles show minor/major; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; eQTL, expression quantitative locus with p<0.05 in EOC tumors; histone marks and DNAse I hypersensitive sites from HaploReg v 4.1 indicating tissue types as defined therein; super enhancer information based on the human super-enhancer database available at http://bioinfo.au.tsinghua.edu.cn/dbsuper/index.php; none of these SNPs had previous GWAS associations with any phenotype based on the EBI GWAS catalog or resided within promoter histone marks; all SNPs are intronic to the gene indicated.
Fig 1. Association of rs2256787 in the ARHGEF10L gene with invasive endometrioid EOC risk by study site and combined.
Squares represent the estimated per-allele odds ratio (OR) and are proportional to sample size for each study; lines indicate its 95% confidence interval (CI); source indicates contributing study;[11] MAF, control minor allele frequency; PVal, per-allele p-value adjusted for age, site, and principal components to account for residual differences in European ancestry.
Three other variants were associated at p-value<10−4 (Table 1, S1, S2 and S3 Figs). rs10788679 in an intron of ARHGEF10L was associated with risk of invasive serous EOC (OR = 1.07, 95% CI: 1.03–1.11, p = 2.6x10-4;); ARHGEF10L SNPs rs2256787 and rs10788679 are independent (r2 = 0.02, 1000 Genomes Project EUR). In addition, rs1955513 was most strongly associated with all invasive EOC risk (OR = 0.90, 95% CI: 0.85–0.95, p = 3.3x10-4). This variant lies in an intron of A-kinase (PRKA) anchor protein 6 (AKAP6). Another variant in AKAP6, intronic SNP rs927062, was also associated with all invasive EOC risk (p = 5.9x10-4); AKAP6SNPs rs1955513 and rs927062 are in modest linkage disequilibrium (r2 = 0.15, 1000 Genomes Project EUR).
We investigated whether the four variants of interest, rs2256787, rs10788679, rs1955513, rs927062, which are all intronic, alter expression of their proximal GTPases, or coincide with regulatory marks that may affect expression (Table 1). In publicly available databases,[13, 14] the ARHGEF10L SNPs rs2256787and rs10788679 coincide with a human ovary super-enhancer, a region of the genome with unusually strong enrichment for the binding of transcriptional coactivators in this tissue. As ARHGEF10L rs2256787 associated with endometrioid EOC risk, we were particularly interested in eQTLs in the 54 endometrioid patients; however, there was no evidence of association between rs2256787 genotype and ARHGEF10L expression in endometrioid EOC tumors or other tumor subtypes. In 312 invasive EOC tumors, the G allele of AKAP6 rs927062 correlated with reduced expression of Rho GTPase activating protein 5 (ARHGAP5), a GTPase ~150kb upstream of AKAP6 (β = -0.22, 95% CI: -0.41 to -0.03, p = 6.6x10-3). Other unstudied variants may also be associated with expression of ARHGAP5 (or may be more strongly associated than rs927062), thus future genome-wide or pathway-based analysis of GTPase SNP-expression relationships are of great interest. In other histology-specific eQTL analyses, none of the four variants tested were associated with EOC tumor mRNA expression.
Conclusion
We investigated 322 SNPs in 88 genes encoding small GTP binding proteins of the Ras superfamily (Ras, Rho, Rab, Ral, Arf, and Ran) in germline DNA of over 17,000 EOC patients and 26,000 controls. The 88 genes were derived from G protein (guanine nucleotide-binding proteins) signaling, Ras-GTPases, regulation of Rho GTPase protein signal transduction and activation of Rac GTPase activity. [18] Ras-GTPases are activated at the plasma membrane by guanine nucleotide exchange factors (GEF) such as: son of sevenless homologs 1 and 2 (Drosophila) (SOS-1 and SOS-2); Ras protein-specific guanine nucleotide-releasing factor 1 (GRF1); Rap guanine nucleotide exchange factor 1 (GRF2); and RasGEF domain family, members 1A, 1B and 1C (RasGRF). They are inactivated by GTPase activating proteins (GAP) which include RAS p21 protein activator (GTPase activating protein) 1 (p120RasGAP). GEF factors are recruited to the plasma membrane by scaffold and adaptor complexes such as SHC/Grb2 that associate with activated tyrosine kinase receptors (TKR).[19] These factors exchange GTP for GDP on the Ras protein. The resulting GTP-Ras protein activates various downstream effectors such as MAP-kinase Raf-1 which activates the MEK/ERK gene regulation cascade, a primary cell growth and anti-apoptosis pathway.[6] Ras-GTPases family members regulate the action of other GTPase pathways involving Rap, Ral, Rac and Rho Ras-GTPase. Ras-GTPases also regulate phosphoinositide 3-kinase (PI3K) and phospholipase C (PLC) activities.[5] Several of these genes are mutated in ovarian tumors.[20]
Overall, analysis at only one SNP yielded a p-value < 10−5: rs2256787 in ARHGEF10L which was associated with 33% increased endometrioid EOC risk. Of note, the experiment-wide error rate for this SNP, accounting for the initial overall set of 6103 candidate SNPs equals 0.027 (Bonferroni-corrected p-value 4.5 x 10−6 x 6103); additionally accounting for six case groups analyzed, this value increases to 0.16 (Bonferroni-corrected p-value 4.5 x 10−6 x 6103 x 6). However, as SNPs, as well as case groups, are not independent, simulation studies are necessary to derive an empirical p-value. Another ARHGEF10L SNP, rs10788679, in showed the smallest p-value in analysis of serous EOC and was the second-most strongly associated SNP in all analyses. ARHGEF10L is a member of the RhoGEF family GEFs that activate Rho GTPases.[21] The Rho branch of the Ras super family encompasses 20 genes in humans, of which Rho, Rac and Cdc42 are the best characterized. Rho GTPases regulate the actin cytoskeleton and control changes in cell morphology and cell motility triggered by extracellular stimuli. Rho GTPases are regulated by GDP/GTP exchange factors and GAPs. Members of this subfamily are activated by specific GEFs and are involved in signal transduction. SNPs in this gene are also associated with obesity[22] and cutaneous basal cell carcinoma.[23]
The SNP most associated with risk of invasive EOC was rs1955513 in the AKAP6 gene. This gene is involved in overall G protein signaling. SNPs in this gene are also associated with neurologic functioning [24] and anorexia.[25] Functionally, rs927062 in AKAP6 was associated with expression of the Rho GTPase activating protein 5, ARHGAP5, also known as p190 RhoGAP, which negatively regulates RHO GTPases. The p190 RhoGAP gene contains a carboxy-terminal domain that functions as a GAP for the Rho family GTPases. In addition to its RhoGAP domain, p190 contains an amino-terminal domain that contains sequence motifs found in all known GTPases.
In conclusion, our study identified potentially functional genetic variants in small GTPase genes that may have roles in EOC susceptibility. To interpret these associations, we suggest consideration of effect sizes and directionality in the context of the sets of histotype-specific analyses conducted; whether a more conservative or liberal statistical significance threshold is applied, the small set of variants highlighted for detailed functional follow-up remain the same. A limitation of this work is that nearby imputed variants were not examined and thus other ungenotyped variants may be driving the reported associations. Nonetheless, four variants in two genes show promising associations that have not been reported previously but point to known pathways that are mutated in ovarian tumors. The results of our investigation suggest that further assessment of this important pathway is warranted in additional collections of densely genotyped EOC patients and controls.
Supporting information
Squares represent the estimated per-allele odds ratio (OR) and are proportional to sample size for each study; lines indicate its 95% confidence interval (CI); Source indicates contributing study [11]; MAF, control minor allele frequency; PVal, per-allele p-value adjusted for age, site, and residual European principal components.
(TIFF)
Squares represent the estimated per-allele odds ratio (OR) and are proportional to sample size for each study; lines indicate its 95% confidence interval (CI); Source indicates contributing study [11]; MAF, control minor allele frequency; PVal, per-allele p-value adjusted for age, site, and residual European principal components.
(TIFF)
Squares represent the estimated per-allele odds ratio (OR) and are proportional to sample size for each study; lines indicate its 95% confidence interval (CI); Source indicates contributing study [11]; MAF, control minor allele frequency; PVal, per-allele p-value adjusted for age, site, and residual European principal components.
(TIFF)
More details are available upon request.
(XLS)
More details are available upon request.
(XLSX)
Acknowledgments
We thank all the individuals who took part in this study and all the researchers, clinicians and technical and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward, P. Webb and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); the staff of the genotyping unit, S LaBoissiere and F Robidoux (Genome Quebec); U. Eilber (GER); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T. W. Park, Simon, K. Beer- Grondke and D. Schmidt (HJO); S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WYL. The authors assume full responsibility for analyses and interpretation of these data (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila, and Y. Bensman (NJO); M. Sherman, A. Hutchinson,N. Szeszenia—‐ Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini,P. Harrington the SEARCH team and ECRIC (SEA); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan and J. Ford (UKO); Carole Pye (UKR); A. Amin Al Olama, K. Michilaidou, K. Kuchenbaker (COGS). The Australian Ovarian Cancer Study acknowledges the cooperation of the participating institutions in Australia and acknowledges the contribution of the study nurses, research assistants and all clinical and scientific collaborators to the study. The complete Australian Ovarian Cancer Study Management Group can be found at www.aocstudy.org (Georgia.Trench@qimrberghofer.edu.au). We would like to thank all of the women who participated in these research programs.
Data Availability
All relevant data are available from the Cambridge research repository Apollo (DOI: https://doi.org/10.17863/CAM.20843).
Funding Statement
The scientific development and funding for this project were funded by the following: NIH R01 CA-1491429 (Phelan PI); the US National Cancer Institute (R01-CA076016); the COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 HEALTH F2 2009-223175); the Genetic Associations and Mechanisms in Oncology (GAME‐ON): a NCI Cancer Post-GWAS Initiative (U19-CA148112); the Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07). Investigator-specific funding: G.C.-T. is supported by the National Health and Medical Research Council; B.K. is funded by the American Cancer Society Early Detection Professorship (SIOP-06‐258‐01‐COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124; L.E.K. is supported by a Canadian Institute of Health Research New Investigator Award (MSH-87734). Funding of included studies: Funding of the constituent studies was provided by the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); the Canadian Institutes of Health Research (MOP-86727); Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124); the Danish Cancer Society (94-222-52); the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy For Cancer Control from the Ministry of Health Labour and Welfare of Japan; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07-CA095666, K07-CA143047, K22-CA138563, N01-CN55424, N01-PC67001, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P01-CA087969, P30-CA072720, P50-CA105009, P50-CA136393, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01- CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA067262, R01-CA071766, R01-CA074850, R01-CA080742, R01-CA080978, R01-CA083918, R01-CA087538, R01-CA092044, R01-095023, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01-CA136924, R03-CA113148, R03-CA115195, U01-CA069417, U01-CA071966, UM1-CA182910, UM1-CA186107 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-07-0449, W81XWH-10-1-02802); the US Public Health Service (PSA-042205); The National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01GB 9401); the State of Baden-Wurttemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS-39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital “Womens Health Theme” and the Royal Marsden Hospital; WorkSafeBC.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Soslow RA. Histologic subtypes of ovarian carcinoma: an overview. Int J Gynecol Pathol. 2008;27(2):161–74. doi: 10.1097/PGP.0b013e31815ea812 [DOI] [PubMed] [Google Scholar]
- 3.Pharoah PD, Ponder BA. The genetics of ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2002;16(4):449–68. [DOI] [PubMed] [Google Scholar]
- 4.Kar SP, Berchuck A, Gayther SA, Goode EL, Moysich KB, Pearce CL, et al. Common genetic variation and susceptibility to ovarian cancer: current insights and future directions. Cancer Epidemiol Biomarkers Prev. 2017. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 5.Pajic M, Herrmann D, Vennin C, Conway JR, Chin VT, Johnsson AK, et al. The dynamics of Rho GTPase signaling and implications for targeting cancer and the tumor microenvironment. Small GTPases. 2015;6(2):123–33. doi: 10.4161/21541248.2014.973749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Just WW, Peranen J. Small GTPases in peroxisome dynamics. Biochim Biophys Acta. 2016;1863(5):1006–13. doi: 10.1016/j.bbamcr.2016.01.004 [DOI] [PubMed] [Google Scholar]
- 7.Li G, Marlin MC. Rab family of GTPases. Methods Mol Biol. 2015;1298:1–15. doi: 10.1007/978-1-4939-2569-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shirakawa R, Horiuchi H. Ral GTPases: crucial mediators of exocytosis and tumourigenesis. J Biochem. 2015;157(5):285–99. doi: 10.1093/jb/mvv029 [DOI] [PubMed] [Google Scholar]
- 9.Nussinov R, Tsai CJ, Chakrabarti M, Jang H. A new view of Ras Isoforms in cancers. Cancer Res. 2016;76(1):18–23. doi: 10.1158/0008-5472.CAN-15-1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41(9):996–1000. doi: 10.1038/ng.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45(4):362–70, 70e1-2. doi: 10.1038/ng.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, et al. STrengthening the REporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. PLoS Med. 2009;6(2):e22 doi: 10.1371/journal.pmed.1000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016;44(D1):D164–71. doi: 10.1093/nar/gkv1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81. doi: 10.1093/nar/gkv1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58(2):362–70. doi: 10.1016/j.molcel.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014;106(10):dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Winterhoff BJ, Kalli KR, Block MS, Armasu SM, Larson MC, et al. Expression signature distinguishing two tumour transcriptome classes associated with progression-free survival among rare histological types of epithelial ovarian cancer. Br J Cancer. 2016;114(12):1412–20. doi: 10.1038/bjc.2016.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoon JL, Tan MH, Koh CG. The regulation of cellular responses to mechanical cues by Rho GTPases. Cells. 2016;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz G, Henninger C. Rho GTPases: novel players in the regulation of the DNA damage response? Biomolecules. 2015;5(4):2417–34. doi: 10.3390/biom5042417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McConechy MK, Ding J, Senz J, Yang W, Melnyk N, Tone AA, et al. Ovarian and endometrial endometrioid carcinomas have distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 2014;27(1):128–34. doi: 10.1038/modpathol.2013.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler S, Mohl M, Wieland T, Lutz S. GrinchGEF—a novel Rho-specific guanine nucleotide exchange factor. Biochem Biophys Res Commun. 2005;335(4):1280–6. doi: 10.1016/j.bbrc.2005.08.025 [DOI] [PubMed] [Google Scholar]
- 22.Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7(12):e51954 doi: 10.1371/journal.pone.0051954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stacey SN, Gudbjartsson DF, Sulem P, Bergthorsson JT, Kumar R, Thorleifsson G, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40(11):1313–8. doi: 10.1038/ng.234 [DOI] [PubMed] [Google Scholar]
- 24.Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53949). Mol Psychiatry. 2015;20(2):183–92. doi: 10.1038/mp.2014.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry. 2011;16(9):949–59. doi: 10.1038/mp.2010.107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Squares represent the estimated per-allele odds ratio (OR) and are proportional to sample size for each study; lines indicate its 95% confidence interval (CI); Source indicates contributing study [11]; MAF, control minor allele frequency; PVal, per-allele p-value adjusted for age, site, and residual European principal components.
(TIFF)
Squares represent the estimated per-allele odds ratio (OR) and are proportional to sample size for each study; lines indicate its 95% confidence interval (CI); Source indicates contributing study [11]; MAF, control minor allele frequency; PVal, per-allele p-value adjusted for age, site, and residual European principal components.
(TIFF)
Squares represent the estimated per-allele odds ratio (OR) and are proportional to sample size for each study; lines indicate its 95% confidence interval (CI); Source indicates contributing study [11]; MAF, control minor allele frequency; PVal, per-allele p-value adjusted for age, site, and residual European principal components.
(TIFF)
More details are available upon request.
(XLS)
More details are available upon request.
(XLSX)
Data Availability Statement
All relevant data are available from the Cambridge research repository Apollo (DOI: https://doi.org/10.17863/CAM.20843).