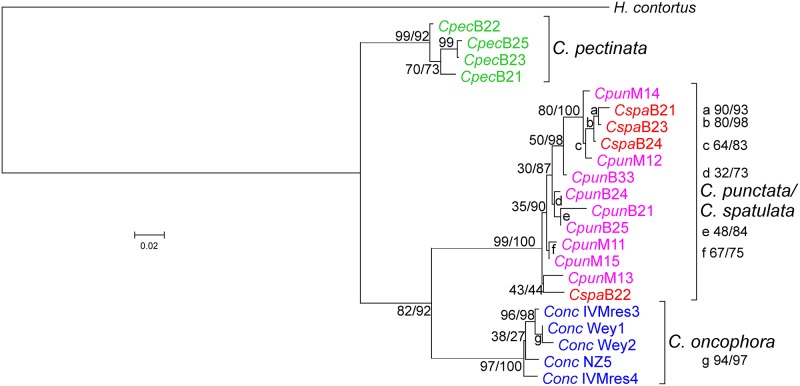
Fig 6. Multi-locus phylogenetic analysis of Cooperia species infecting cattle.
Sequences were aligned using M-Coffee (partial isotype 1 β-tubulin and mitochondrial cox-2 genes) or MAFFT (partial ITS-1, complete 5.8S, ITS-2 fragment and partial mitochondrial 12S rRNA genes). Protein coding regions were manually edited to ensure that codons were not disrupted by gaps. A phylogenetic tree was calculated using RAxML with one partition per gene except for the cox-2 gene for which separate partitions for codon positions 1 and 2 and codon position 3 were included. Sequences from Haemonchus contortus (accession numbers DQ469245 + KT428386 + EU346694were used as outgroup. Samples were obtained from individual worms identified as Cooperia pectinata (Cpec, green), Cooperia punctata (Cpun, magenta) from Brazil, and Cooperia spatulata (Cspa, red) and from pools of larvae from Cooperia oncophora (Conc, blue) and C. punctata from Mexico. Sequences derived from Brazil (B) and Mexico (M) are indicated together with numbers indicating the particular voucher (in combination with Cooperia morphospecies and geographical origin. The C. oncophora and the Mexican C. punctata samples were obtained from different pools of larvae using isolates that have been characterized as single species isolates. Node support values represent results of the rapid bootstrapping analysis before and of the Shimodaira-Hasegawa likelihood ratio test behind the slash. Accession numbers for all new sequences are available from S2 Table.