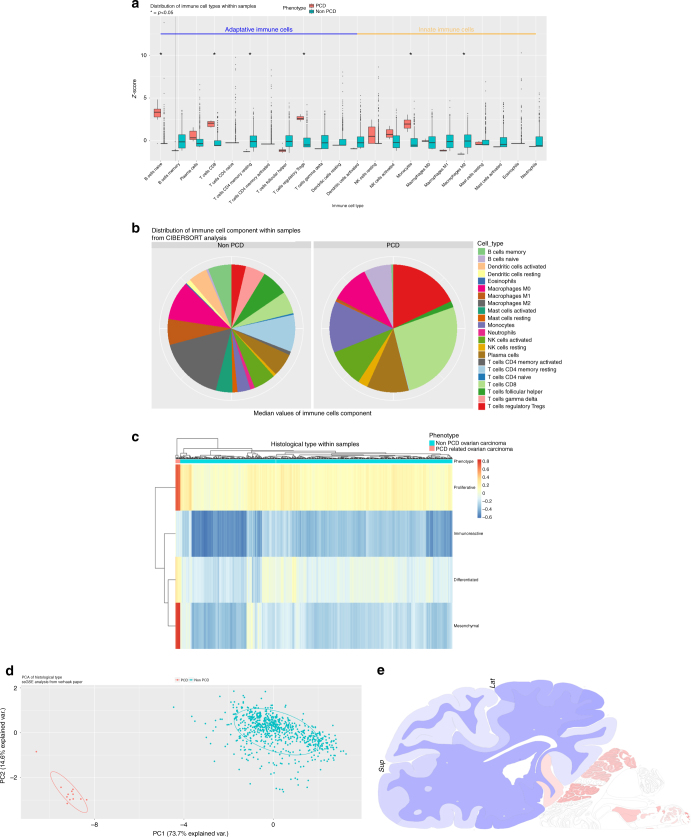
Fig. 4.
a Boxplots with z-score of the distribution of the different cell types between PCD and non-PCD related ovarian tumour samples. Analysis was performed with CIBERSORT algorithm. Classification regarding adaptive immune system (on the left) and innate immune system (on the right) was made. Statistical significance is shown by “*”, and significant cells are written in bold. b pie chart representing the distribution of the immune cell types (mean values) in non-PCD samples (on the left) and PCD samples (on the right). Data from CIBERSORT analysis. c Heatmap of the histological subtypes scores. Annotation on columns correspond to the samples' phenotypes (PCD or non-PCD samples). Data from single sample gene set enrichment analysis on gene sets from the literature(20). Hierarchical clustering was used to cluster the samples. d Principal component analysis (PCA) was the histological subtypes results matrix. Plot using first and second PCA (explaining respectively 73.7 and 14.6% of variance) and coloured membership of PCD samples (blue) and non-PCD samples (orange). e: heatmap of CDR1, CDR2 and CDR2L genes expression across the brain. Data from the brainscope website using Allen brain institute project data. Brain is shown from coronal view through the cerebellum, with a 90° left rotation. Colour legend indicates expression value going from blue to red scale