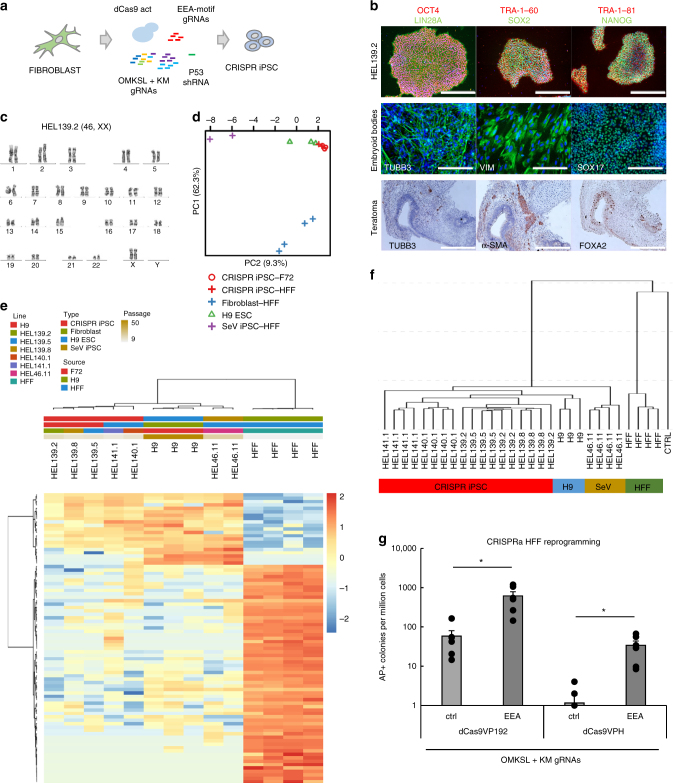
Fig. 3.
EEA-motif targeting enhances derivation of CRISPRa iPSCs from primary skin fibroblasts. a Schematic representation of skin fibroblast reprogramming with dCas9 activators. b Pluripotency factor expression in CRISPR-iPSC colonies (top row, scale bar = 400 µm) and tri-lineage differentiation markers for ectoderm (TUBB3), mesoderm (Vimentin and α-SMA), and endoderm (SOX17 and FOXA2) in embryoid bodies (middle row, scale bar = 200 µm) and teratomas (bottom row, scale bar = 800 µm). c Normal 46, XX karyotype of a CRISPRa iPSC line HEL139.2. d Principal component analysis of CRISPR iPSC lines, control PSC lines and HFFs based on expression of 123 significantly fluctuated genes. e Clustering of iPSC lines and HFFs based on expression of 85 significantly fluctuated and differentially regulated genes. f Clustering of CRISPR iPSC lines and control pluripotent stem cells based on DNA methylation. g Effect of VP192 and VPH domains and EEA-motif targeting on CRISPRa reprogramming efficiency of HFFs. n = 6 from three independent experiments. Data presented as mean ± s.e.m., two tailed Student’s t-test. **P < 0.01