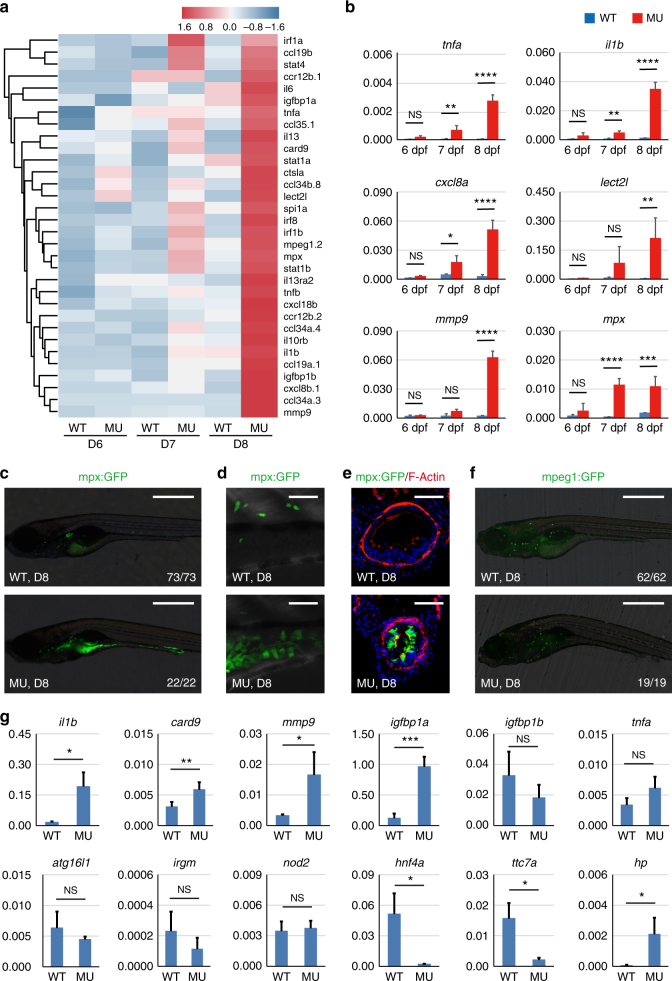
Fig. 3.
Pro-inflammatory responses in the intestine of pik3c3 mutants. a Heatmap of inflammation-related genes shows the gradual stimulation of inflammatory response from 6 to 8 dpf in the mutants. b qRT-PCR analysis of the indicated inflammation-related genes. cxcl8a is the zebrafish homolog of IL8. Experiments were performed and data analyzed as described in Fig. 2b. c mpx:GFP is a transgenic zebrafish line with neutrophils labeled by GFP. In 8 dpf mutants, neutrophils mainly accumulate in the digestive tract. Scale bar, 500 µm. d High magnification confocal imaging of (c). Scale bar, 50 µm. e Cross sections of intestinal bulb in D8 embryos. Neutrophil infiltration of intestinal epithelium is clearly seen in the mutants. Scale bar, 50 µm. f mpeg1:GFP-labeled macrophage lineage cells appear normal in the mutants. Scale bar, 500 µm. g qRT-PCR analysis of the expression of IBD candidate genes in dissected intestine of pik3c3 mutants at 8 dpf. Inflammatory response genes (il1b, card9, mmp9) and igfbp1a (one of the homologs of human IGFBP1, a gene related to the prognosis of CD) are significantly up-regulated in the mutants. Bacteria sensing and autophagy-related genes (nod2, atg16l1, and irgm) and tnfa are not stimulated. Barrier function-related genes hnf4a and ttc7a are severely down-regulated in the mutants while the hp gene, which functions to open up tight junctions, is up-regulated in the mutants. actb1 is used as internal control and data represent mean ± SD from three biological repeats. p-values are determined by unpaired two-tailed Student’s t-test (NS, non-significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001)