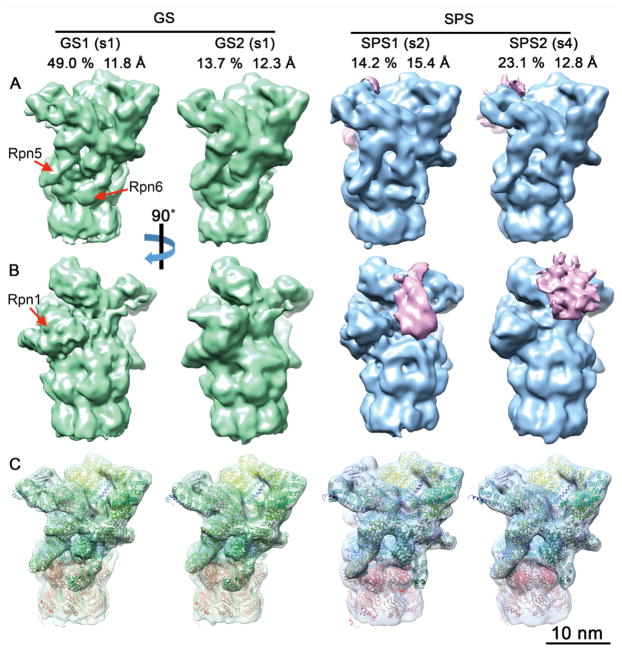
Figure 3. Subtomogram Classification of 26S Proteasomes Reveals Enrichment of Substrate Processing Conformations.
To analyze the functional state of proteasome regulatory particles single- and double-capped proteasomes were cut by the half of the CP. The resulting half proteasomes were classified according to RP conformation into ground or substrate processing states (Asano et al., 2015), yielding two ground states (GS1, GS2) and two substrate processing classes (SPS1, SPS2). (A, B) The four density maps are displayed in solid surface representation in two different views. The positions of the Rpn1, Rpn5 and Rpn6 subunits are indicated. Prominent densities in the substrate binding region of SPS1 and SPS2 are colored in pink. For each class, the percentage of the total number of classified particles and the global resolution are indicated. (C) Same view as (A), with semi-transparent maps superimposed with the atomic models generated by MDFF. The classes respectively represent the s1 state with different Rpn1 positions (GS1, GS2), the s2 state (SPS1) and the s4 state (SPS2). Atomic models are colored by subunits: Rpn1 (brown), Rpn2 (yellow), Rpn9/5/6/7/3/12 (different shades of green), Rpn8/Rpn11 (light/dark magenta), Rpn10 and Rpn13 621 (purple), AAA-ATPase hexamer (blue), and CP (red). See also Figure S2B, Figure S3 and Table S1.