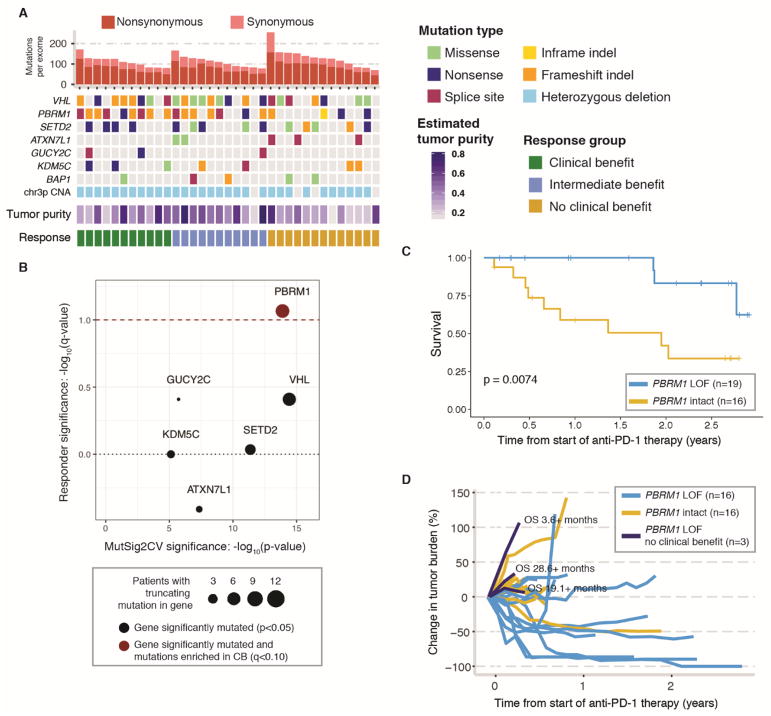
Fig. 2. Analysis of tumor genome features in discovery cohort reveals a correlation between PBRM1 LOF mutations and clinical benefit from anti-PD-1 therapy.
(A) Mutations in the discovery cohort. Patients are ordered by response category, with tumor mutation burden in decreasing order within each response category. Shown are the genes that were recurrently mutated at a significant frequency, as assessed by MutSig2CV analysis (table S1E). CNA = copy number alteration. (B) Enrichment of truncating mutations in tumors from patients in the CB vs. NCB groups. Red dashed line denotes q<0.1 (Fisher’s exact test). Mutations in genes above the black dotted line are enriched in tumors of patients with CB from anti-PD-1 therapy and mutations in genes below the line are enriched in tumors of patients with NCB. (C) Kaplan-Meier curve comparing overall survival of patients treated with anti-PD-1 therapy whose tumors did or did not harbor LOF mutations in PBRM1. See also fig. S5 for Kaplan-Meier curve comparing progression-free survival of these patients. (D) Spider plot showing objective decrease in tumor burden in PBRM1-LOF (blue) vs. PBRM1-intact (yellow) tumors. Three patients with early progression on anti-PD-1 therapy and truncating mutations in PBRM1 (dark blue) had long and/or censored OS.