Table 1.
Estimates of genetic diversity within each subpopulation.
| Breeds | Nsamples | Estimated on all markers |
Estimated on independent markers |
||||
|---|---|---|---|---|---|---|---|
| Nall_M1 | Ppoly2 | PH-W3 | Nindep_M4 | Lr2 = 0.25 |
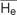 6 6
|
||
| Chahua | 97 | 46211 | 84.66% | 99.27% | 5980 | 7.5 | 0.299 |
| Silkie | 100 | 80.31% | 99.45% | 4726 | 72 | 0.333 | |
| Langshan | 96 | 84.08% | 99.36% | 4926 | 62.5 | 0.309 | |
| Beard | 95 | 94.82% | 99.27% | 13225 | 3.5 | 0.362 | |
1Nall_M indicates means the number of all markers after quality control.
2Ppoly indicates the percentage of polymorphic loci, with minor allele frequencies more than 0.05.
3PH-W indicates the percentage of SNP loci conformed to Hardy–Weinberg expectations.
4Nindep_M indicates the number of independent markers in each subpopulation.
5Lr2 = 0.2 indicates the average physical distance (Kb) between two adjacent markers when their r2 drop to 0.2.
6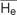 indicates the average expected heterozygosity.
indicates the average expected heterozygosity.
