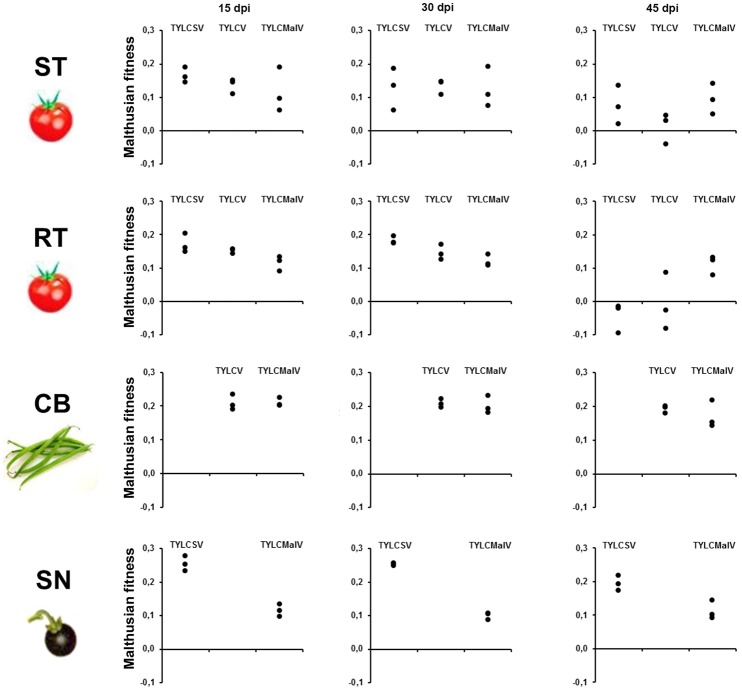
Figure 3.
Absolute fitness of begomovirus during time-course analyses of tomato yellow leaf curl Sardinia virus (TYLCSV), tomato yellow leaf curl virus (TYLCV) and tomato yellow leaf curl Malaga virus (TYLCMaV) infections in susceptible tomato (ST), resistant tomato (RT), common bean (CB) and the wild reservoir Solanum nigrum (SN). Absolute fitness of begomovirus DNA molecules present in the apical leaves of plants infected with infectious begomovirus clones was approximated by calculating the Malthusian growth rate per day, m, calculated according to the formula logQ, where Q is the number of pg of begomovirus ssDNA per 100 ng of total plant DNA. Three individual systemically infected plants (as judged by hybridization analysis) for each virus/host combination were monitored. Apical leaf samples (non-inoculated) were taken at 15, 30 and 45 days post inoculation (dpi). No viral detection was possible at any time point for TYLCSV/common bean or TYLCV/S. nigrum infections either by hybridization or qPCR.