Abstract
F-box proteins are essential components of the Skp-cullin-F-box complex (a type of E3 ubiquitin ligase), and participate in cell cycle and immune responses through the ubiquitin proteasome system. F-box protein 39 (FBXO39) belongs to the F-box family, which has been reported to be associated with cancer oncogenesis and progression. The present study aimed to investigate the role of FBXO39 in osteosarcoma (OS) cell proliferation and apoptosis in vitro. It was demonstrated that U-2OS cells exhibited high expression of FBXO39 compared with HOS and SaOS-2 osteosarcoma cells. Thus, knockdown of FBXO39 was performed using lentivirus-mediated short hairpin RNA (shRNA) transfection to validate the effect of FBXO39 in U-2OS cells. Western blotting and RT-qPCR analysis were used to confirm the efficiency of infection by analyzing the expression level of FBXO39. Using Celigo-based cell counting and MTT assays, it was demonstrated that FBXO39 knockdown significantly reduced the rate of cell proliferation compared with control. Caspase 3/7 activity assays and fluorescence-activated cell sorting confirmed the induction of apoptosis in U-2OS cells following FBXO39 knockdown. In conclusion, it was demonstrated that FBXO39 knockdown may significantly inhibit proliferation and promote apoptosis of U-2OS cells. Thus, FBXO39 may serve an important role in OS progression.
Keywords: F-box protein 39, osteosarcoma, U-2OS cells, proliferation, apoptosis
Introduction
Osteosarcoma (OS) is the most common type of primary malignant bone tumor in children and young adults, originating from mesenchymal cells and is characterized by the production of immature osteoid (1,2). Due to the combination of surgery and multi-agent chemotherapy, the 5-year survival rate of non-metastatic OS has risen from 25 to 60% in the last decade (3). However, >20% patients with OS exhibit lung metastases at initial diagnosis. Lung metastasis occurs following surgery in ~80% patients with OS (4,5). Therefore, the 5-year survival rate for metastatic OS is only 30%, a figure which has remained constant for 30 years (6). Previous studies reported a variety of genetic alterations in OS, however, the molecular mechanism underlying OS development remains to be elucidated (7–9). Therefore, there is an urgent requirement to characterize the molecular mechanisms underlying OS development in order to identify novel and effective targets for OS chemotherapy.
The Skp1-cullin-F-box (SCF) complex is a member of the E3 ubiquitin ligase family, involved in substrate recognition, ubiquitination recruitment and degradation in the ubiquitin proteasome system (10). The F-box family proteins belong to a critical subunit of the SCF complex, characterized by their ~40 amino-acid motifs. F-box proteins recognize and combine substrates in the SCF complex, and participate in a series of cellular processes, including cell cycle and immune responses (10–13). It has been reported that F-box protein 39 (FBXO39) was highly expressed in normal human testis and abnormally expressed in a variety of cancer cell types (14,15). Furthermore, it has been demonstrated that patients with colon cancer exhibit a high level of serum FBXO39 expression compared with healthy individuals (14,15).
In the present study, it was demonstrated that FBXO39 was highly expressed in OS, and that FBXO39 knockdown inhibited cell proliferation and promoted apoptosis in human osteosarcoma U-2OS cells.
Materials and methods
Cell culture
The human OS cell lines, HOS, SaOS-2 and U-2OS, were purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Corning Incorporated, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS; Vian-saga, Shanghai, China) at 37°C in a humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from HOS, Saos-2 and U-2OS cells using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Reverse transcription was performed using M-MLV Reverse Transcriptase, RNase Inhibitor and dNTPs (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. RT-qPCR was performed using SYBR Master mix (Takara Biotechnology Co., Ltd., Dalian) and a Roche Light Cycler 480 Real-time PCR system (Roche Diagnostics, Basel, Switzerland). GAPDH was used as an internal control gene. Quantification was performed using the 2−ΔΔCq method (16). The primer sequences were as follows: FBXO395 forward, 5′-GATGGGCAAACGCCTGGATTA-3′ and reverse, 5′-GGAGGGTGCTGGCATTCTCAC-3′; GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′, and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. The thermal cycle included: 95°C for 15 sec, 45 cycles of 95°C for 5 sec, and 60°C for 30 sec.
Lentivirus-mediated short hairpin RNA (shRNA) FBXO39 knockdown
The lentivirus-mediated shRNA vector system was designed, constructed, packed and purified by Shanghai GeneChem Co, Ltd. (Shanghai, China), and all procedures were performed according to the manufacturer's protocol. Cells transfected with lentivirus containing human FBXO39 shRNA were the experimental group, denoted as shFBXO39 group in the subsequent experiments. Cells transfected with lentivirus containing blank shRNA were used as a negative control, denoted as shCtrl group. The multiplicity of infection was 10 (2×105 cells transfected per well and 2×106 TU lentivirus transfected per well in a 6-well plate), and the infection was proceeded with the addition of DMEM and 5 µg/ml Polybrene (Clontech Laboratories, Inc., Mountainview, CA, USA) to the cells. The fluorescent microscopy (Olympus IX71; Olympus Corporation, Tokyo, Japan) demonstrated that the transfection efficiency of U-2OS cells transfected with shFBXO39 and shCtrl lentivirus was >80%. The knockdown efficiency of the target gene was detected by RT-qPCR and western blotting 3 days after transfection.
Western blot analysis
Western blotting was used to validate lentiviral knockdown efficiency at the protein level in U-2OS cells. Proteins were extracted from cells using a lysis buffer (2% 2-mercaptoethanol, 4% SDS and 20% glycerol, 100 mM Tris-HCl), and the protein concentration was measured using a bicinchoninic acid (BCA) Protein Assay kit (Beyotime Institute of Biotechnology, Haimen, China). A total of 20 µg protein was separated by SDS-PAGE (10% gels), and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The plasmid was constructed with a flag epitope, and GAPDH was used as a loading control. The membranes were blocked in Tris-buffered saline with Tween (TBST) containing 5% non-fat milk overnight at 4°C. The membranes were then incubated with the following primary antibodies: Flag (dilution, 1:2,000; cat. no. F1804; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and GAPDH (dilution, 1:2,000; cat. no. sc-32233; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. The membranes were then washed three times in TBST. The membranes were incubated with a horseradish peroxidase-conjugated goat anti-mouse immunoglobulin secondary antibody (dilution, 1:2,000; cat. no. sc-2005; Santa-Cruz Biotechnology, Inc.) for 2 h at room temperature, then washed three times in TBST. Bands were visualized using enhanced chemiluminescence (ECL; Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Celigo analysis
U-2OS cells were seeded at a density of 2×103 cells/well in a 96-well plate at 72 h post-transfection (at 37°C in an atmosphere of 5% CO2). After plating, Celigo® Image Cytometer (Nexcelom, Lawrence, MA, USA) was used to evaluate the number of cells by scanning green fluorescence daily for 5 days at room temperature (17,18).
MTT assay
MTT assay was used to analyze the effect of FBXO39 knockdown on cell proliferation (19). Cells transfected with shCtrl and shFBXO39 were seeded in 96-well plates at a density of 2×103 cells/well at 72 h post-transfection. A total of 20 µl 5 mg/ml MTT was added to each well for 4 h at 37°C. Then, the medium was discarded and 110 µl dimethyl sulfoxide (Shanghai Test Chemical Reagent Co, Ltd., Shanghai. China) was added to each well to dissolve the formazan crystals. The optical density (OD) was detected at 490 nm using M2009PR Multifunctional Microplate Reader (Tecan Group, Ltd., Mannedorf, Switzerland).
Caspase 3/7 activity analysis
Cells transfected with shCtrl or shFBXO39 were seeded in 96-well plates and incubated at 37°C for 5 days. Caspase-Glo reaction solution was made up by mixing 10 ml Caspase-Glo 3/7 buffer solution and Caspase-Glo 3/7 substrate (Caspase-Glo® 3/7 Assay; G8091; Promega Corporation, Madison, WI, USA). A total of 100 µl Caspase-Glo reaction solution was added per well, each well containing 1×104 cells. Following a 2-h incubation at room temperature, a microplate reader was used to detect the luminescence intensity at 570 nm.
FACS analysis
Fluorescence-activated cell sorting (FACS) was used to analyze cell apoptosis (20). Cells transfected with shCtrl or shFBXO39 were plated in 6-cm dishes 5 days after transfection and grown to 70% confluence. Following washing with binding buffer [from the Annexin V-allophycocyanin (APC) Detection kit (cat. no. 88-8007; eBioscience; Thermo Fisher Scientific, Inc.] once, cells were stained with 200 µl binding buffer containing 10 µl APC Detection kit for 10–15 min at room temperature in the dark. A flow cytometer (Merck KGaA) and InCyte 3.1 (Merck KGaA) were then used to analyze the cells.
Statistical analysis
Statistical analysis was performed using SPSS 19.0 software (IBM Corp, Armonk, NY, USA). All data are presented as the mean of ≥3 independent experiments. The error bars represent standard deviation. Differences between 2 groups were determined using Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
U-2OS cells express a high level of FBXO39 compared with HOS and SaOS-2 cells
The expression level of FBX039 mRNA was analyzed in OS cells lines using RT-qPCR (Fig. 1). It was demonstrated that the U-2OS cell line exhibited the highest expression of FBXO39 among the three cell lines. Based on this result, U-2OS cells were selected for subsequent experiments.
Figure 1.
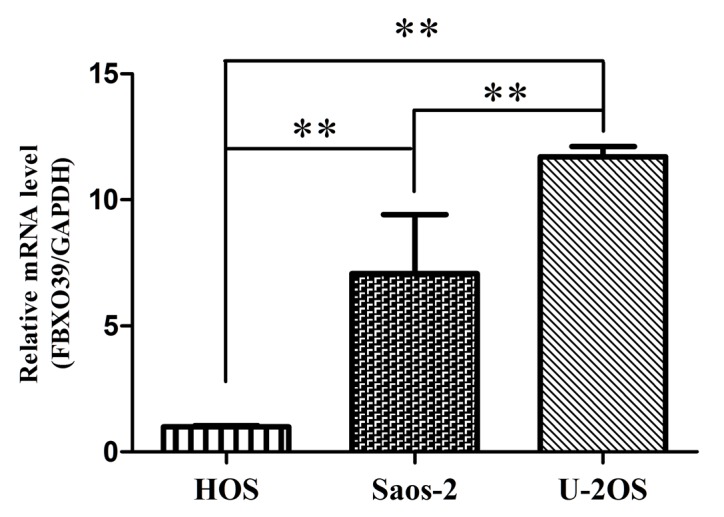
The relative mRNA expression level of FBXO39 in human osteosarcoma cell lines, HOS, SaOS-2 and U-2OS, was measured by reverse transcription-quantitative polymerase chain reaction. **P<0.01. FBXO39, F-box protein 39.
FBXO39 knockdown in U-2OS cells by lentiviral transfection
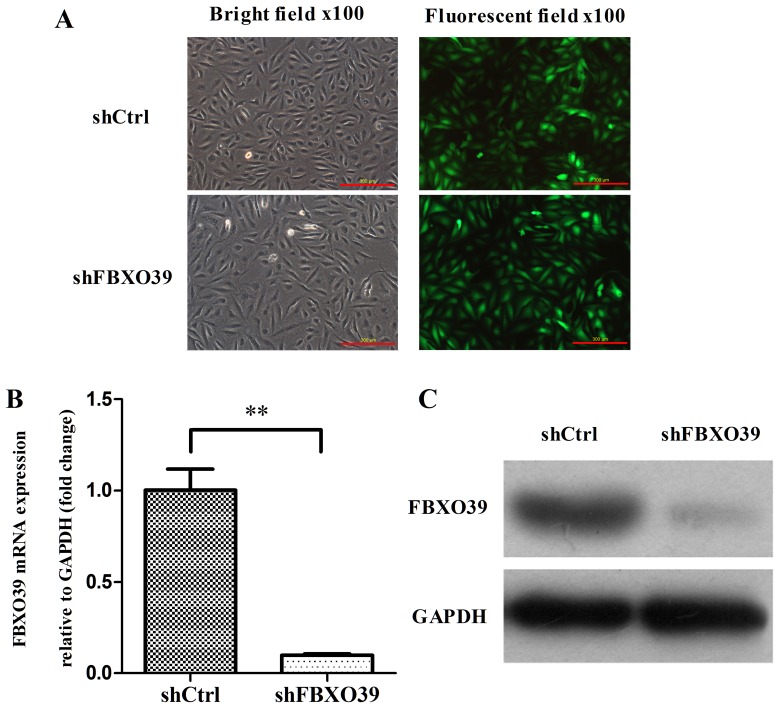
To investigate the role of FBXO39 on OS cell proliferation and apoptosis, FBXO39 was knocked down using lentiviral vectors. Transfection efficiency was assessed by fluorescence imaging 72 h after transfection (Fig. 2A). According to the results, >80% cells transfected with lentivirus expressed green fluorescent protein (data not shown). RT-qPCR and western blotting analyses were conducted to assess the knockdown efficiency on the mRNA and protein levels, respectively (Fig. 2A-C). The mRNA expression level of FBXO39 in U-2OS cells transfected with shFBXO39 was significantly decreased compared with that in cells transfected with shCtrl (P<0.01). Similarly, the FBXO39 protein expression level was decreased in U-2OS cells transfected with shFBXO39 compared with cells transfected with shCtrl (Fig. 2C). Thus, FBX039 knockdown reduced FBXO39 expression at the mRNA and protein level.
Figure 2.
FBXO39 knockdown using shRNA in U-2OS cells. (A) Transfection efficiency was examined by fluorescence imaging. (B) FBXO39 mRNA expression was determined by reverse transcription-quantitative polymerase chain reaction. **P<0.01. (C) FBXO39 protein expression level was detected by western blot analysis in U-2OS cells. FBXO39, F-box protein 39; shRNA, short hairpin RNA; shFBXO39, cells transfected with FBXO39-targeting shRNA; shCtrl, cells transfected with control shRNA.
FBXO39 knockdown inhibits the proliferation of U-2OS cells
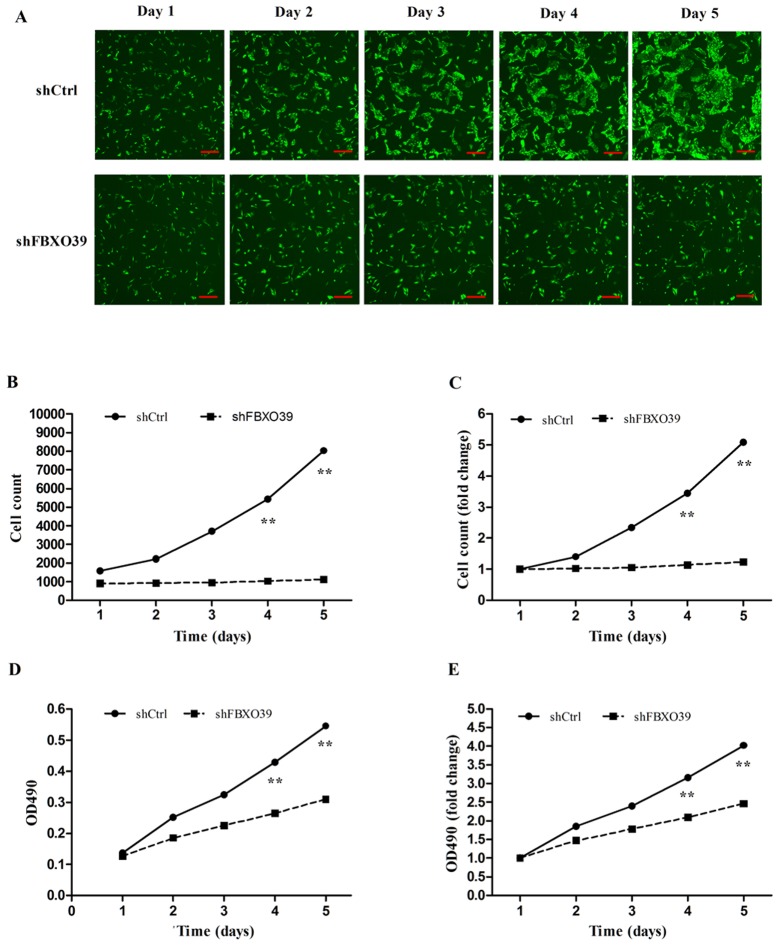
To investigate the effect of FBXO39 knockdown on cell proliferation in U-2OS cells, Celigo analysis and MTT assays were performed. Celigo analysis revealed that the proliferation of cells transfected with shFBOX39 was significantly inhibited 5 days after transfection compared with control (P<0.01; Fig. 3A-C). On day 5, the cell count of cells transfected with shCtrl was 5.08-fold greater compared with that on day 1, whereas the cell count of cells transfected with shFBXO39 only increased by 1.23-fold (Fig. 3C). The MTT assay also demonstrated that FBXO39 knockdown inhibited proliferation compared with control (P<0.01; Fig. 3D and E). In conclusion, FBXO39 knockdown inhibited proliferation of U-2OS cells.
Figure 3.
FBXO39 knockdown inhibits the proliferation of U-2OS. (A) Celigo analysis of shCtrl and shFBXO39 cells was performed 3 days after transfection. The increasing of fluorescence (cell numbers) in shFBXO39 cells was significantly inhibited. All images were captured at the original magnification. Scale bars, 300 µm. (B) Graph presenting the cell numbers over 5 days. (C) Graph presenting the cell growth rate, obtained by comparing the cell count of each day to that of day 1. (D) The OD490 was detected by MTT assay each day for 5 days after transfection. OD490 indicated the amount of viable cells. The proliferation of shFBXO39 cells was significantly inhibited. (E) The fold change of OD490 (the cell growth rate) was obtained by comparing the OD490 of each day to that of day 1. The cell growth rate of shFBXO39 was notably lower. **P<0.01, vs. shCtrl. FBXO39, F-box protein 39; shFBXO39, cells transfected with FBXO39-targeting shRNA; shCtrl, cells transfected with control shRNA; OD490, optical density at 490 nm.
FBXO39 knockdown promotes apoptosis of U-2OS cells
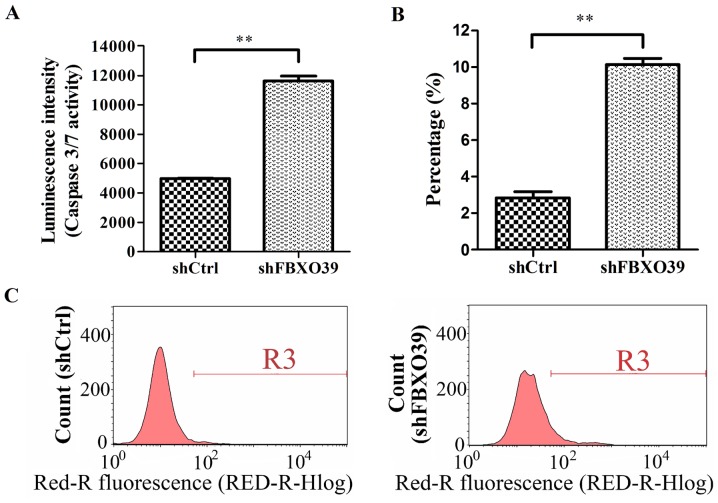
To investigate the effect of FBXO39 knockdown on U-2OS cell apoptosis, cells transfected with shFBXO39 and shCtrl were subjected to FACS analysis and caspase 3/7 activity analysis (Fig. 4A-C). FACS analysis revealed a significant increase in the apoptotic rate in shFBXO39 cells compared with shCtrl cells 5 days after transfection (P<0.01; Fig. 4B and C). Cell apoptosis was further analyzed by detecting activated caspase 3/7. In this assay, it was demonstrated that cells transfected with shFBXO39 exhibited an increased level of activated caspase 3/7 activity compared with that in cells transfected with shCtrl (P<0.01; Fig. 4A). In conclusion, FBXO39 knockdown promoted apoptosis in U-2OS cells.
Figure 4.
FBXO39 knockdown promotes apoptosis of U-2OS cells. (A) Caspase 3/7 activity analysis was performed in cells transfected with shCtrl and shFBXO39. The luminescence intensity indicated Caspase 3/7 activity. (B) The percentage of apoptotic cells was detected by FACS in each group. The result represented the mean of three repetitively independent assays. (C) Representative histograms of the FACS results. Apoptotic cells were stained with Annexin V and detected by flow cytometry. **P<0.01. FBXO39, F-box protein 39; shFBXO39, cells transfected with FBXO39-targeting shRNA; shCtrl, cells transfected with control shRNA; FACS, fluorescence-activated cell sorting.
Discussion
Despite efforts to improve the diagnosis and therapy in patients with OS, the 5-year survival rate and prognosis of patients with OS remains poor (9). An improved understanding of OS progression is required for the development of novel therapies. Previous studies have reported several genes associated with OS, including MYC, FBJ murine osteosarcoma viral oncogene homolog, Mouse double minute 2, RECQ helicase, tumor protein P53, retinoblastoma 1, kruppel like factor 8 and collagen triple helix repeat containing 1 (21–24).
F-box proteins serve a key role in the regulation of cellular functions, including proliferation, apoptosis and immune responses, by detecting and recruiting substrates for ubiquitination (10). F-box proteins are associated with the carcinogenesis and progression of various types of cancer. For example, S-phase kinase associated protein 2 has been demonstrated to be highly expressed in cancer cells, and its downregulation may promote oncogenesis (25). In contrast, F-box and WD repeat domain containing 7 (FBXW7) has been demonstrated to function as a tumor-suppressor in several types of cancer, and upregulation of FBXW7 has been indicated to reduce cancer cell proliferation and migration (26–28). FBXO39 has been demonstrated to be highly expressed in normal testis tissue and several types of cancer (14,15). The association between OS and FBXO39 was explored in U-2OS cells in the present study.
The results of the present study revealed that U-2OS cells exhibited the highest expression level of FBXO39 among OS cell lines. Additionally, a significant decrease in proliferation was observed in response to FBXO39 downregulation compared with control as assessed by Celigo and MTT analysis. These results suggest that FBXO39 may promote proliferation in OS cells. Furthermore, FACS and caspase 3/7 analysis revealed that FBXO39 knockdown in U-2OS cells caused an increase in the rate of apoptosis compared with control. These results demonstrate that FBXO39 knockdown inhibited proliferation and promoted apoptosis of human OS cells.
In conclusion, the present study indicates that FBXO39 serves an important role in OS carcinogenesis and progression. Future studies that examine cellular function, including migration and invasion, as well as signaling pathways and in vivo experiments are required to elucidate the role of FBXO39 in OS.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Sanming Project of Medicine in Shenzhen; The National Natural Science Foundation of China (grant no. 21602137) and the Shenzhen Health and Family Planning Science Project (grant no. 201501014).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SZ and JZ conceived and designed the study. CZ, PW, JC, XJ, ZZhu, ZZha and AG performed the experiments, and collected and interpreted the data. SZ, JZ, WY, WL and JT analyzed the data and edited the draft manuscript. TJ and WG analyzed the data and revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 2.Heare T, Hensley MA, Dell'Orfano S. Bone tumors: Osteosarcoma and Ewing's sarcoma. Curr Opin Pediatr. 2009;21:365–372. doi: 10.1097/MOP.0b013e32832b1111. [DOI] [PubMed] [Google Scholar]
- 3.Caudill JS, Arndt CA. Diagnosis and management of bone malignancy in adolescence. Adolesc Med State Art Rev. 2007;18:62–78. ix. [PubMed] [Google Scholar]
- 4.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9:422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 5.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 6.Ando K, Heymann MF, Stresing V, Mori K, Rédini F, Heymann D. Current therapeutic strategies and novel approaches in osteosarcoma. Cancers (Basel) 2013;5:591–616. doi: 10.3390/cancers5020591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge JA, Nelson M, McComb E, McGuire MH, Rosenthal H, Vergara G, Maale GE, Spanier S, Neff JR. Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer Genet Cytogenet. 1997;95:74–87. doi: 10.1016/S0165-4608(96)00306-8. [DOI] [PubMed] [Google Scholar]
- 8.Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer. 2003;3:685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- 9.Basu-Roy U, Basilico C, Mansukhani A. Perspectives on cancer stem cells in osteosarcoma. Cancer Lett. 2013;338:158–167. doi: 10.1016/j.canlet.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews3002. REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho MS, Ou C, Chan YR, Chien CT, Pi H. The utility F-box for protein destruction. Cell Mol Life Sci. 2008;65:1977–2000. doi: 10.1007/s00018-008-7592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uddin S, Bhat AA, Krishnankutty R, Mir F, Kulinski M, Mohammad RM. Involvement of F-BOX proteins in progression and development of human malignancies. Semin Cancer Biol. 2016;36:18–32. doi: 10.1016/j.semcancer.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Song MH, Ha JC, Lee SM, Park YM, Lee SY. Identification of BCP-20 (FBXO39) as a cancer/testis antigen from colon cancer patients by SEREX. Biochem Biophys Res Commun. 2011;408:195–201. doi: 10.1016/j.bbrc.2011.02.077. [DOI] [PubMed] [Google Scholar]
- 15.Seifi-Alan M, Shamsi R, Ghafouri-Fard S, Mirfakhraie R, Zare-Abdollahi D, Movafagh A, Modarressi MH, Kazemi G, Geranpayeh L, Najafi-Ashtiani M. Expression analysis of two cancer-testis genes, FBXO39 and TDRD4, in breast cancer tissues and cell lines. Asian Pac J Cancer Prev. 2014;14:6625–6629. doi: 10.7314/APJCP.2013.14.11.6625. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Nabzdyk CS, Chun M, Pradhan L, Logerfo FW. High throughput RNAi assay optimization using adherent cell cytometry. J Transl Med. 2011;9:48. doi: 10.1186/1479-5876-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, Hardisson D, Eccles SA. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moodley S, Koorbanally NA, Moodley T, Ramjugernath D, Pillay M. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay is a rapid, cheap, screening test for the in vitro anti-tuberculous activity of chalcones. J Microbiol Methods. 2014;104:72–78. doi: 10.1016/j.mimet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Pan Y, Shan W, Fang H, Guo M, Nie Z, Huang Y, Yao S. Sensitive and visible detection of apoptotic cells on Annexin-V modified substrate using aminophenylboronic acid modified gold nanoparticles (APBA-GNPs) labeling. Biosens Bioelectron. 2014;52:62–68. doi: 10.1016/j.bios.2013.07.057. [DOI] [PubMed] [Google Scholar]
- 21.Smida J, Baumhoer D, Rosemann M, Walch A, Bielack S, Poremba C, Remberger K, Korsching E, Scheurlen W, Dierkes C, et al. Genomic alterations and allelic imbalances are strong prognostic predictors in osteosarcoma. Clin Cancer Res. 2010;16:4256–4267. doi: 10.1158/1078-0432.CCR-10-0284. [DOI] [PubMed] [Google Scholar]
- 22.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin F, Shen Z, Tang LN, Zheng SE, Sun YJ, Min DL, Yao Y. KLF8 knockdown suppresses proliferation and invasion in human osteosarcoma cells. Mol Med Rep. 2014;9:1613–1617. doi: 10.3892/mmr.2014.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sang W, Zhu L, Ma J, Lu H, Wang C. Lentivirus-mediated knockdown of CTHRC1 inhibits osteosarcoma cell proliferation and migration. Cancer Biother Radiopharm. 2016;31:91–98. doi: 10.1089/cbr.2014.1758. [DOI] [PubMed] [Google Scholar]
- 25.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welcker M, Clurman BE. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 27.Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J, Delrow JJ, Small T, Grady WM, Nakayama KI, Clurman BE. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol Cell Biol. 2012;32:2160–2167. doi: 10.1128/MCB.00305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: The role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.