Abstract
MicroRNAs (miRs) are an emerging class of non-coding, endogenous and small RNA molecules that serve important functions in tumorigenesis and development. The present study investigated the expression, functions and molecular mechanism underlying miR-205 in hepatocellular carcinoma. miR-205 was downregulated in hepatocellular carcinoma tissues and cell lines. Ectopic miR-205 expression suppressed hepatocellular carcinoma cell proliferation, migration and invasion in vitro. In addition, vascular endothelial growth factor A (VEGFA) was identified as a functional downstream target of miR-205 in hepatocellular carcinoma. Furthermore, knockdown of VEGFA revealed the same functions with miR-205 overexpression in hepatocellular carcinoma cells. These results provided evidence that miR-205 served important functions in the inhibition of hepatocellular carcinoma cells growth and metastasis via directly targeting VEGFA, which indicated that miR-205 may have therapeutic value for hepatocellular carcinoma.
Keywords: hepatocellular carcinoma, vascular endothelial growth factor A, microRNA-205, metastasis, growth
Introduction
Hepatocellular carcinoma (HCC), a major subtype of primary liver cancer, represents the sixth most prevalent malignancy and the third most common cause of cancer-associated mortalities globally (1,2). According to statistics, in 2015, 35,660 new HCC cases and 24,550 mortalities due to HCC were predicted in the USA (3). HCC initiation and progression involves a variety of risk factors, including chronic infection with hepatitis C virus or hepatitis B virus (HBV), alcohol or drug abuse and a high-fat or high-sugar diet (4). Among these risk factors, infection with HBV is the most prevalent cause of the disease worldwide, and is responsible for the increasing incidence of HCC (1,4). Developments have been made in improving diagnosis and therapeutic treatments for HCC, but due to difficulties in diagnosis, a high frequency of recurrence and resistance to common chemotherapy and radiotherapy agents, the prognosis for patients with HCC remains poor (5–7). The 5-year survival rate is <5% for patients with HCC and intra-hepatic or extra-hepatic metastasis (8). Therefore, it is urgent to elucidate the molecular mechanisms underlying HCC carcinogenesis and progression, and to develop effective strategies for the diagnosis, treatment and prognosis of HCC.
Previously, numerous studies have suggested that the abnormal expression of microRNAs (miRNAs/miRs) is associated with the carcinogenesis, progression, metastasis and recurrence of HCC (9,10). miRNAs are an emerging class of non-coding, endogenous, small RNA molecules, ~19–25 nucleotides in length, that negatively regulate the protein production of target mRNAs at the post-transcriptional level by interacting with the 3′untranslated regions (3′UTR) of target mRNAs (11,12). In this manner, miRNAs were involved in various biological processes, including differentiation, proliferation, angiogenesis, metabolism, apoptosis, cell cycle and metastasis (13,14). Accumulated evidence has suggested that miRNAs may act as a novel group of oncogenes or tumor suppressors, and the deregulation of miRNA and target mRNA expression levels may contribute to carcinogenesis and cancer development in a substantial number of human malignancies, including HCC (15–17). Therefore, further investigation concerning the expression levels, functions and molecular mechanisms underlying the effects of miRNAs in HCC will provide insight into the carcinogenesis and progression of HCC.
The aim of the present study was to investigate the expression levels, functions and molecular mechanisms underlying the effect of miR-205 in HCC. Initially, the expression levels of miR-205 in HCC tissues and cell lines were evaluated. Secondly, the effects of miR-205 on the proliferation and metastasis of HCC cells were determined by MTT assay, migration and invasion assays. Finally, vascular endothelial growth factor A (VEGFA) was identified as a functional downstream target of miR-205 in HCC. These results indicated that miR-205 may function as a tumor suppressor in HCC by directly targeting VEGFA, suggesting a potential targeted therapy for patients with HCC.
Materials and methods
Tissue specimens, cell culture and cell transfection
The present study was approved by the Ethics Committee of Guizhou Cancer Hospital (Guiyang, China). Written informed consent was obtained from all patients with HCC prior to enrollment in the present study. None of the patients had received antitumor treatment prior to surgery. HCC tissues and matched adjacent non-cancerous liver tissues were obtained from 32 patients (19 male; 13 female; age range, 46–74 years; mean age, 62 years) who underwent surgery at Guizhou Cancer Hospital between August 2013 and January 2015. All tissue specimens were immediately frozen and stored in liquid nitrogen.
HepG2, HuH-7, SMMC-7721 and BEL-7402 human HCC cell lines and L02 and HEK293T normal hepatic cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 0.1 mg/ml streptomycin (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C with 5% CO2.
An miR-205 mimic and a negative control (NC) were purchased from Genepharm, Inc. (Sunnyvale, CA, USA); the miR-205 mimic sequence was 5′-UCCUUCAUUCCACCGGAGUCUG-3′ and the NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. VEGFA small interfering (si)RNA and NC siRNA were also purchased from Genepharm, Inc. The sequence for VEGFA siRNA and NC siRNA were as follows: VEGFA siRNA sense, 5′-GGCAGAAUCAUCACGAAGUTT-3′ and antisense, 5′-ACUUCGUGAUGAUUCUGCCTT-3′; NC siRNA sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT−3′. miRNA and siRNA transfection was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues (1 g) or cells (2×106) was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A TaqMan miRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for quantification of miR-205 expression levels, according to the manufacturer's protocol. For quantification of VEGFA mRNA expression levels, reverse transcription was performed using the M-MLV Reverse Transcription system (Promega Corporation, Madison, WI, USA). The temperature protocol for reverse transcription was as follows: 95°C for 2 min; 20 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 2 min; then 72°C for 5 min. qPCR was performed using the SYBR-Green Master Mix (Takara Biotechnology Co., Ltd., Dalian, China). The thermocycling conditions for qPCR were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. U6 and β-actin were used as internal controls for miR-205 and VEGFA expression levels, respectively. The primers were as follows: miR-205, 5′-GCTCCTTCATTCCACCGG-3′ (forward) and 5′-CAGTGCAGGGTCCGAGGT-3′ (reverse); U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); VEGFA, 5′-AACTTTCTGCTGTCTTGGGT-3′ (forward) and 5′-TCTCGATTGGATGGCAGTA-3′ (reverse); and β-actin, 5′-GGGACCTGACTGACTACCTC-3′ (forward) and 5′-TCATACTCCTGCTTGCTGAT-3′ (reverse). Each sample was analyzed in triplicate. Relative expression levels were calculated using the 2−∆∆Cq method (18).
MTT assay
The cell proliferation rate was evaluated using the MTT assay (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). HepG2 and HuH-7 cells were seeded into 96-well plates at a density of 3,000 cells per well. Following overnight incubation, miRNA or siRNA transfection was performed as aforementioned. MTT assays were performed following transfection at room temperature for 24, 48, 72 and 96 h. In brief, 20 µl MTT solution was added to each well. Following incubation with MTT solution at 37°C for an additional 4 h, 200 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to dissolve the formazan at 37°C for 10 min. Finally, the absorbance at 490 nm was detected using an ELISA reader (BioTek Instruments, Inc., Winooski, VT, USA). Each sample was analyzed in triplicate.
Cell migration and invasion assays
Cell migration and invasion were evaluated using Transwell chambers (Corning Incorporated, Corning, NY, USA) with 8 µm pore-size polycarbonate membranes. For the invasion assay, Transwell chambers were pre-coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). For the migration and invasion assays, 3×104 transfected HepG2 and HuH-7 cells suspended in 300 µl serum-free DMEM were added to the upper chamber. DMEM (500 µl) supplemented with 20% FBS was added to the low chamber as a chemoattractant. Following incubation at 37°C for 48 h, cells that had not migrated or invaded to the bottom surface of the Transwell chambers were carefully removed with cotton wool. The migrated and invaded cells were fixed with 100% methanol at room temperature for 10 min, stained with 0.5% crystal violet for 10 min and imaged with a light microscope (magnification, ×200).
Western blotting
Total protein was isolated from HepG2 and HuH-7 cells (2×106) using RIPA lysis buffer (Bioteke Corporation, Beijing, China) supplemented with 0.1 mg/ml phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate and 1 mg/ml aprotinin. Protein concentration was determined using a bicinchoninic acid assay kit (Bioteke Instuments, Inc.). Total protein (20 µg) was fractionated by 10% SDS-PAGE (Beyotime Institute of Biotechnology, Haimen, China), followed by transference to polyvinylidene (PVDF) membranes (Millipore, Bedford, MA). Non-specific binding sites of PVDF membranes were blocked using 10% non-fat milk in Tris-buffered saline with 0.1% Tween (TBST) solution at room temperature for 1 h. Subsequently, the membranes were probed with mouse anti-human VEGFA monoclonal primary antibody (dilution, 1:1,000; cat. no., ab155944;) and mouse anti-human GADPH monoclonal primary antibody (dilution, 1:1,000; cat. no., ab9484; both Abcam, Cambridge, UK). Following incubation overnight at 4°C, membranes were washed with TBST three times and incubated with goat anti mouse IgG horseradish peroxidase-conjugated secondary antibody (1:3,000 dilution; ab6789; Abcam) for 2 h at room temperature. Following washing three times with TBST, the membranes were visualized using enhanced chemiluminescence solution (Pierce; Thermo Fisher Scientific, Inc.).
Dual-luciferase reporter assay
The target genes of miR-205 were assessed using the miRNA target prediction tool TargetScan (version 6.0; http://www.targetscan.org/vert_60/) (19). In order to explore whether VEGFA was a direct target gene of miR-205, a dual-luciferase reporter assay was performed. HEK293T cells were co-transfected with VEGFA-3′UTR wild-type (Wt) or VEGFA-3′UTR mutant (Mut), miR-205 mimics or NC using Lipofectamine 2000 according to the manufacturer's protocol. Luciferase activities were evaluated 48 h following transfection using a Dual-Luciferase Reporter System (Promega Corporation, Madison, WI, USA). Renilla luciferase activity was detected as an internal control for firefly luciferase activity. Each sample was analyzed in triplicate.
Statistical analysis
Data were presented as the mean ± standard deviation. Statistical analyses were performed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). All statistical analyses were two-tailed. Data were analyzed using Student's t-tests and one-way analysis of variance, with Student-Newman-Keuls tests used to compare two groups in analyses with multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-205 expression level was decreased in HCC tissues and cell lines
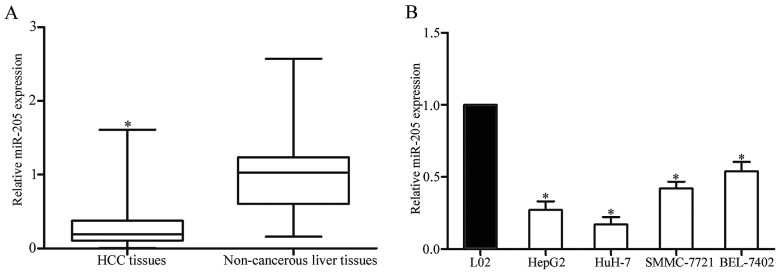
In order to determine miR-205 expression levels and its function in HCC, expression levels of miR-205 were detected in HCC tissues and matched adjacent non-cancerous liver tissues using RT-qPCR. As presented in Fig. 1A, miR-205 was significantly downregulated in HCC tissues compared with in matched adjacent non-cancerous liver tissues. In addition, miR-205 expression levels in HCC cell lines and a normal hepatic cell line were also determined. As presented in Fig. 1B, the expression levels of miR-205 were decreased in the four HCC cell lines compared with the L02 normal hepatic cell line. These results indicated that the downregulation of miR-205 may serve an important function in HCC carcinogenesis and progression.
Figure 1.
miR-205 expression levels were decreased in HCC tissues and cell lines. (A) Expression levels of miR-205 in HCC tissues and matched adjacent non-cancerous liver tissues were determined by reverse transcription-quantitative polymerase chain reaction. (B) Expression levels of miR-205 in HepG2, HuH-7, SMMC-7721 and BEL-7402 HCC cell lines and the L02 normal hepatic cell line. *P<0.05 vs. respective controls. HCC, hepatocellular carcinoma; miR, microRNA.
miR-205 inhibited the proliferation of HCC cells
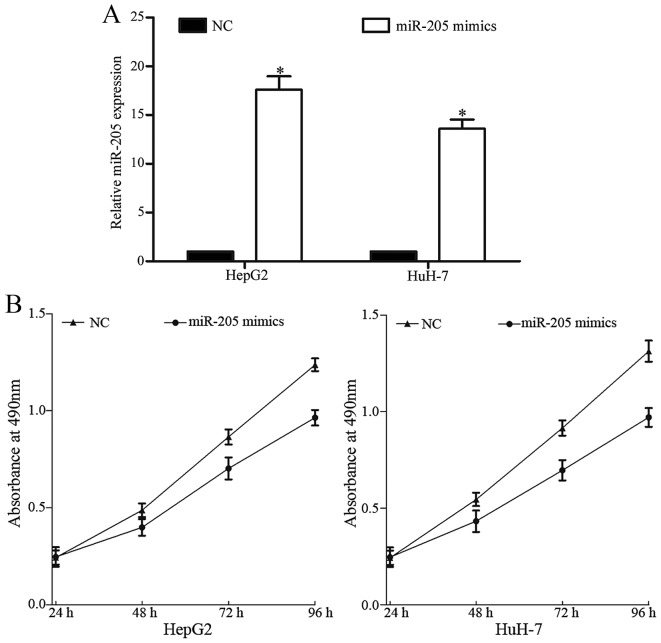
In order to investigate the functions of miR-205 in HCC, miR-205 mimics or NC were transfected into HepG2 and HuH-7 cells. As presented in Fig. 2A, miR-205 was upregulated in HepG2 and HuH-7 cells following transfection with miR-205 mimics compared with NC, suggesting that HepG2 and HuH-7 cells were effective and adjustable models for the functional studies of miR-205.
Figure 2.
miR-205 inhibited the proliferation of HCC cells. (A) Expression levels of miR-205 in HepG2 and HuH-7 cells following transfection with miR-205 mimics or NC. (B) MTT assays were performed to determine the effect of miR-205 on the proliferation of HepG2 and HuH-7 cells. *P<0.05 vs. NC. HCC, hepatocellular carcinoma. miR, microRNA; NC, negative control.
Cell proliferation was evaluated using the MTT assay. miR-205 significantly reduced HepG2 and HuH-7 cell proliferation (Fig. 2B). These results indicated that overexpression of miR-205 inhibited the proliferation of HepG2 and HuH-7 cells.
MiR-205 inhibited migration and invasion of HCC cells
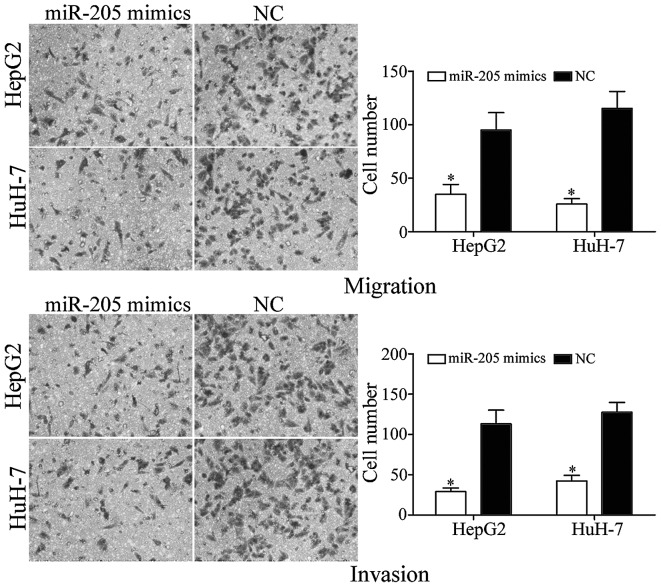
In order to investigate the function of miR-205 in metastasis, Transwell migration and invasion assays were performed. As presented in Fig. 3, upregulation of miR-205 induced the suppression of tumor cell migration and invasion in HepG2 and HuH-7 cells compared with NC controls. These results indicated that miR-205 suppressed HCC cell metastasis in vitro.
Figure 3.
miR-205 overexpression suppressed the migration and invasion of HepG2 and HuH-7 cells (magnification, ×200). *P<0.05 vs. NC. miR, microRNA; NC, negative control.
miR-205 directly targets the 3′UTR of VEGFA
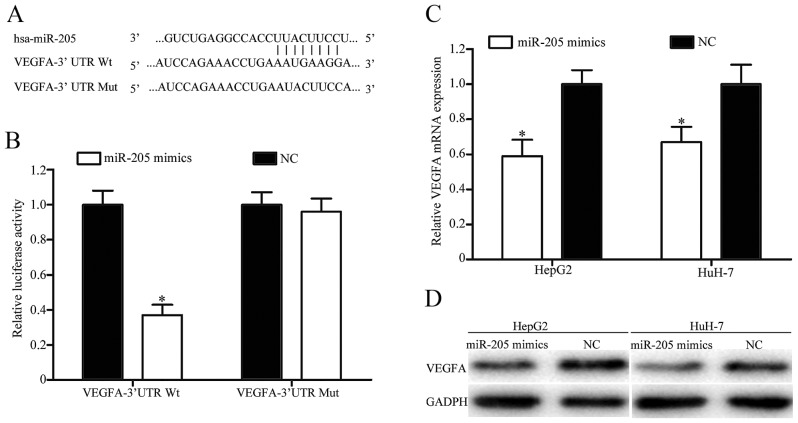
In order to explore the molecular mechanism underlying the function of miR-205 in HCC, TargetScan was used to predict the potential target genes of miR-205. As presented in Fig. 4A, VEGFA was identified to possess a putative miR-205 binding site in the 3′UTR. To verify whether VEGFA was a direct target gene of miR-205, a dual-luciferase reporter assay was performed. The results revealed that overexpression of miR-205 decreased the luciferase activity of VEGFA-3′UTR Wt, but did not decrease the luciferase activity of VEGFA-3′UTR Mut (Fig. 4B). Furthermore, RT-qPCR and western blotting was performed to investigate the effect of miR-205 on the expression levels of VEGFA mRNA and protein. As presented in Fig. 4C and D, upregulation of miR-205 significantly inhibited the expression level of VEGFA in HepG2 and HuH-7 cells at the mRNA and protein levels, respectively. Taken together, these results indicated that VEGFA was a direct target of miR-205 in HCC.
Figure 4.
miR-205 directly targeted VEGFA by binding to its 3′UTR in hepatocellular carcinoma. (A) The predicted miR-205 binding site within VEGFA 3′UTR and its mutated version by site mutagenesis are presented. (B) Dual-luciferase reporter assays were performed to confirm the direct regulation of miR-205 in the 3′UTR of VEGFA. miR-205 decreased VEGFA expression at the (C) mRNA and (D) protein levels in HepG2 and HuH-7 cells. *P<0.05 vs. NC. miR, microRNA; VEGFA, vascular endothelial growth factor A; 3′UTR, 3′untranslated region; NC, negative control; Wt, wild-type; Mut, mutant.
VEGFA is involved in miR-205-induced suppression of HCC cell proliferation, migration and invasion
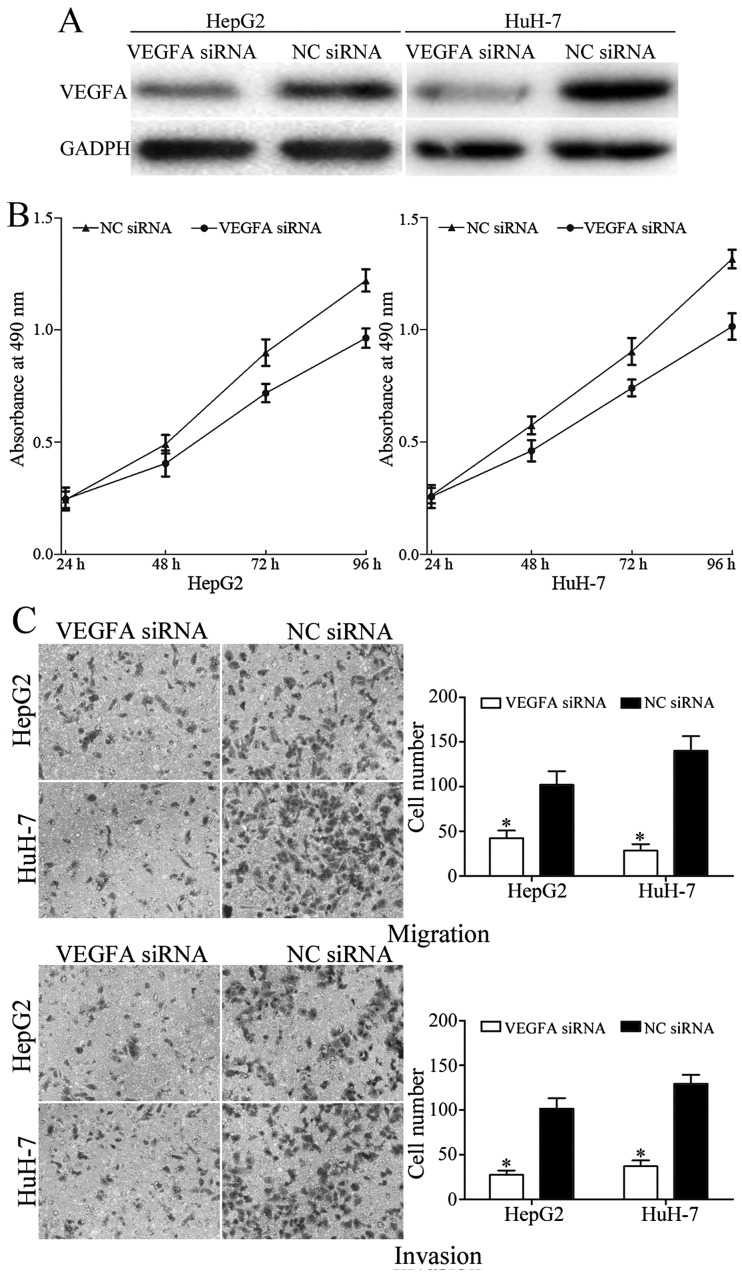
To explore whether VEGFA acted as a critical mediator of miR-205 in HCC, VEGFA siRNA was used to knock down VEGFA expression. As presented in Fig. 5A, VEGFA was significantly downregulated in HepG2 and HuH-7 cells following transfection with VEGFA siRNA. Furthermore, MTT, migration and invasion assays were performed to evaluated the effect of VEGFA siRNA on the growth and metastasis of HCC. The results demonstrated that knockdown of VEGFA suppressed the proliferation, migration and invasion of HepG2 and HuH-7 cells (Fig. 5B and C). Together, these results suggested that miR-205 suppressed growth, migration and invasion of HCC by downregulating VEGFA.
Figure 5.
VEGFA was involved in miR-205-induced suppression of hepatocellular carcinoma proliferation, migration and invasion. (A) Protein expression levels of VEGFA in HepG2 and HuH-7 cells following transfection with VEGFA siRNA or NC siRNA. (B) MTT assays were performed to determine the effect of VEGFA siRNA on the proliferation of HepG2 and HuH-7 cells. (C) Knockdown of VEGFA suppressed migration and invasion of HepG2 and HuH-7 cells (magnification, ×200). *P<0.05 vs. NC. VEGFA, vascular endothelial growth factor A; miR, microRNA; siRNA, short interfering RNA; NC, negative control.
Discussion
It is now widely accepted that miRNAs modulate various biological processes, including cancer initiation and progression. In the present study, miR-205 expression levels in HCC tissues and cell lines were determined, and the biological functions and molecular mechanisms underlying miR-205 in HCC carcinogenesis and progression were also investigated. It was revealed that miR-205 was downregulated in HCC tissues and cell lines. Overexpression of miR-205 significantly suppressed the proliferation, migration and invasion of HCC cells by directly targeting VEGFA. These findings suggested that miR-205 may be a notable tumor suppressor in HCC.
Previous studies have demonstrated that miR-205 functions as an oncogene in numerous types of human cancer (20–22). For example, in laryngeal squamous cell carcinoma, miR-205 was upregulated and enhanced cell growth and invasion via negative regulation of CDK2AP1 expression level (20). Expression levels of miR-205 were reported to be increased in endometrial cancer tissues (21). Kaplan-Meier survival analysis revealed that high expression levels of miR-205 were significantly associated with poor prognosis of patients with endometrial cancer (21). In addition, miR-205 promoted proliferation, metastasis and inhibited apoptosis of endometrial cancer cells by targeting the protein kinase B signaling pathway, phosphatase and tensin homolog (PTEN) and estrogen-related receptor γ (23–25). In ovarian cancer, miR-205 was upregulated and miR-205 expression levels were associated with high pathological grade and advanced clinical stage of patients with epithelial ovarian cancer. Ectopic miR-205 expression improved migration and invasion abilities of ovarian cancer cells by directly targeting zing-finger E-box binding homeobox 1 (22). Lei et al (24) and Bai et al (25) investigated miR-205 expression levels in non-small cell lung cancer (NSCLC) tissues and cell lines. Upregulation of miR-205 increased the proliferation, migration, invasion and chemoresistance of NSCLC cells via regulation of the PTEN signaling pathway (26,27). These results suggested that miR-205 may serve important functions in these types of cancer, and should be investigated as a potential therapeutic target for possible therapeutic strategies.
miR-205 has been reported to be a tumor suppressor in various types of cancer, including osteosarcoma (28), thyroid cancer (29), breast cancer (30,31), renal cell carcinoma (32), oral carcinoma (33) and prostate cancer (34). In accordance with these results, the present study indicated that miR-205 was downregulated in HCC tissues and overexpression of miR-205 inhibited HCC cell proliferation, migration and invasion in vitro. These conflicting results concerning miR-205 expression levels and functions demonstrate that miR-205 acted as an oncogene in certain types of cancer and as a tumor suppressor in others. This contradiction may be explained by the ‘imperfect complementarity’ of the interactions between miRNAs and target genes (35).
In order to understand the molecular mechanisms underlying miR-205-induced suppression of cell proliferation, migration and invasion in HCC, TargetScan and dual-luciferase reporter assays were performed. VEGFA was verified to be a direct target gene of miR-205 in HCC. Furthermore, RT-qPCR and western blotting were preformed to investigate whether miR-205 regulated VEGFA mRNA and protein expression levels. The results revealed that miR-205 significantly decreased VEGFA expression at the mRNA and protein levels. Taken together, miR-205 negatively regulated VEGFA expression by directly binding to the 3′UTR of VEGFA in HCC. Identification of the target gene of miR-205 is essential for understanding its functions in HCC carcinogenesis and progression. It is also important for developing novel therapeutic targets for HCC.
VEGFA, a 35–45 kD heparin-binding glycoprotein, is a key regulator of angiogenesis which is known to be a fundamental factor in the local growth of tumors and progression to metastasis (28). High expression levels of VEGFA have been reported in various types of human cancers, including HCC (36,37). VEGFA serves an important function in tumor proliferation, migration, invasion and angiogenesis (38–40). Therefore, anti-VEGFA targeted therapy, including bevacizumab, has been widely used to treat cancers in a clinical setting (41). Regarding its cancer-associated functions in HCC, VEGFA is worth paying attention to as a potential target to treat patients with HCC. The present study revealed that miR-205 targeted VEGFA to suppress cell proliferation and metastasis in HCC. Collectively, miR-205/VEGFA based targeted therapy may be a novel therapeutic treatment for HCC.
In conclusion, the present study demonstrated that miR-205 was significantly downregulated in HCC tissues and cell lines, and overexpression of miR-205 inhibited the proliferation, migration and invasion of HCC cells by directly targeting VEGFA. These results help to further the understanding of the molecular mechanisms underlying HCC carcinogenesis and progression, and may potentially lead to novel targeted therapies for HCC in future studies.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 6.Huang X, Qin J, Lu S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int J Clin Exp Pathol. 2015;8:1515–1524. [PMC free article] [PubMed] [Google Scholar]
- 7.Yang LY, Fang F, Ou DP, Wu W, Zeng ZJ, Wu F. Solitary large hepatocellular carcinoma: A specific subtype of hepatocellular carcinoma with good outcome after hepatic resection. Ann Surg. 2009;249:118–123. doi: 10.1097/SLA.0b013e3181904988. [DOI] [PubMed] [Google Scholar]
- 8.Zhou YM, Zhang XF, Yu F, Liu XB, Wu LP, Li B, Yang JM. Efficacy of surgical resection for pulmonary metastases from hepatocellular carcinoma. Med Sci Monit. 2014;20:1544–1549. doi: 10.12659/MSM.890853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Bo L, Zhao X, Chen Q. MicroRNA-133a inhibits cell proliferation, colony formation ability, migration and invasion by targeting matrix metallopeptidase 9 in hepatocellular carcinoma. Mol Med Rep. 2015;11:3900–3907. doi: 10.3892/mmr.2015.3232. [DOI] [PubMed] [Google Scholar]
- 10.Zhang ZQ, Meng H, Wang N, Liang LN, Liu LN, Lu SM, Luan Y. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn Pathol. 2014;9:135. doi: 10.1186/1746-1596-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 15.Li B, Liu L, Li X, Wu L. miR-503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem Biophys Res Commun. 2015;464:982–987. doi: 10.1016/j.bbrc.2015.06.169. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Fang L, Yu W, Wang Y. MicroRNA-125b suppresses the migration and invasion of hepatocellular carcinoma cells by targeting transcriptional coactivator with PDZ-binding motif. Oncol Lett. 2015;9:1971–1975. doi: 10.3892/ol.2015.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng C, Li J, Wang Q, Liu W, Zhou J, Liu R, Zeng Q, Peng X, Huang C, Cao P, Cao K. MicroRNA-195 functions as a tumor suppressor by inhibiting CBX4 in hepatocellular carcinoma. Oncol Rep. 2015;33:1115–1122. doi: 10.3892/or.2015.3734. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Zhong G, Xiong X. miR-205 promotes proliferation and invasion of laryngeal squamous cell carcinoma by suppressing CDK2AP1 expression. Biol Res. 2015;48:60. doi: 10.1186/s40659-015-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karaayvaz M, Zhang C, Liang S, Shroyer KR, Ju J. Prognostic significance of miR-205 in endometrial cancer. PLoS One. 2012;7:e35158. doi: 10.1371/journal.pone.0035158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu K, Shen W, Zhang Y, Zhao Y, Lu Y. MiR-205 promotes motility of ovarian cancer cells via targeting ZEB1. Gene. 2015;574:330–336. doi: 10.1016/j.gene.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Jin C, Liang R. miR-205 promotes epithelial-mesenchymal transition by targeting AKT signaling in endometrial cancer cells. J Obstet Gynaecol Res. 2015;41:1653–1660. doi: 10.1111/jog.12756. [DOI] [PubMed] [Google Scholar]
- 24.Su N, Qiu H, Chen Y, Yang T, Yan Q, Wan X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol Rep. 2013;29:2297–2302. doi: 10.3892/or.2013.2400. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G, Hou X, Li Y, Zhao M. MiR-205 inhibits cell apoptosis by targeting phosphatase and tensin homolog deleted on chromosome ten in endometrial cancer Ishikawa cells. BMC Cancer. 2014;14:440. doi: 10.1186/1471-2407-14-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei L, Huang Y, Gong W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep. 2013;30:2897–2902. doi: 10.3892/or.2013.2755. [DOI] [PubMed] [Google Scholar]
- 27.Bai J, Zhu X, Ma J, Wang W. miR-205 regulates A549 cells proliferation by targeting PTEN. Int J Clin Exp Pathol. 2015;8:1175–1183. [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Shan M, Liu Y, Yang F, Qi H, Zhou L, Qiu L, Li Y. miR-205 suppresses the proliferative and migratory capacity of human osteosarcoma Mg-63 cells by targeting VEGFA. Onco Targets Ther. 2015;8:2635–2642. doi: 10.2147/OTT.S80088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salajegheh A, Vosgha H, Rahman Md A, Amin M, Smith RA, Lam AK. Modulatory role of miR-205 in angiogenesis and progression of thyroid cancer. J Mol Endocrinol. 2015;55:183–196. doi: 10.1530/JME-15-0182. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Fan Q. MicroRNA-205 inhibits the proliferation and invasion of breast cancer by regulating AMOT expression. Oncol Rep. 2015;34:2163–2170. doi: 10.3892/or.2015.4148. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Li B, Zhao H, Chang J. The expression and clinical significance of serum miR-205 for breast cancer and its role in detection of human cancers. Int J Clin Exp Med. 2015;8:3034–3043. [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Tang ZY, He Y, Liu LF, Li DJ, Chen X. miRNA-205 is a candidate tumor suppressor that targets ZEB2 in renal cell carcinoma. Oncol Res Treat. 2014;37:658–664. doi: 10.1159/000368792. [DOI] [PubMed] [Google Scholar]
- 33.Kim JS, Park SY, Lee SA, Park MG, Yu SK, Lee MH, Park MR, Kim SG, Oh JS, Lee SY, et al. MicroRNA-205 suppresses the oral carcinoma oncogenic activity via down-regulation of Axin-2 in KB human oral cancer cell. Mol Cell Biochem. 2014;387:71–79. doi: 10.1007/s11010-013-1872-7. [DOI] [PubMed] [Google Scholar]
- 34.Wang N, Li Q, Feng NH, Cheng G, Guan ZL, Wang Y, Qin C, Yin CJ, Hua LX. miR-205 is frequently downregulated in prostate cancer and acts as a tumor suppressor by inhibiting tumor growth. Asian J Androl. 2013;15:735–741. doi: 10.1038/aja.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y, Yu W, Wu X, Ye J, Yang S, et al. Identification of miR-7 as an oncogene in renal cell carcinoma. J Mol Histol. 2013;44:669–677. doi: 10.1007/s10735-013-9516-5. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi R, Yano H, Iemura A, Ogasawara S, Haramaki M, Kojiro M. Expression of vascular endothelial growth factor in human hepatocellular carcinoma. Hepatology. 1998;28:68–77. doi: 10.1002/hep.510280111. [DOI] [PubMed] [Google Scholar]
- 37.Miura H, Miyazaki T, Kuroda M, Oka T, Machinami R, Kodama T, Shibuya M, Makuuchi M, Yazaki Y, Ohnishi S. Increased expression of vascular endothelial growth factor in human hepatocellular carcinoma. J Hepatol. 1997;27:854–861. doi: 10.1016/S0168-8278(97)80323-6. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang Y, Wei M. Impact of vascular endothelial growth factor expression on overall survival in patients with osteosarcoma: A meta-analysis. Tumour Biol. 2014;35:1745–1749. doi: 10.1007/s13277-014-1692-8. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Zheng Q, Wu H, Guo X, Li J, Hao S. Rapamycin increases pCREB, Bcl-2, and VEGF-A through ERK under normoxia. Acta Biochim Biophys Sin (Shanghai) 2013;45:259–267. doi: 10.1093/abbs/gmt002. [DOI] [PubMed] [Google Scholar]
- 40.Wiszniak S, Mackenzie FE, Anderson P, Kabbara S, Ruhrberg C, Schwarz Q. Neural crest cell-derived VEGF promotes embryonic jaw extension. Proc Natl Acad Sci USA. 2015;112:6086–6091. doi: 10.1073/pnas.1419368112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]