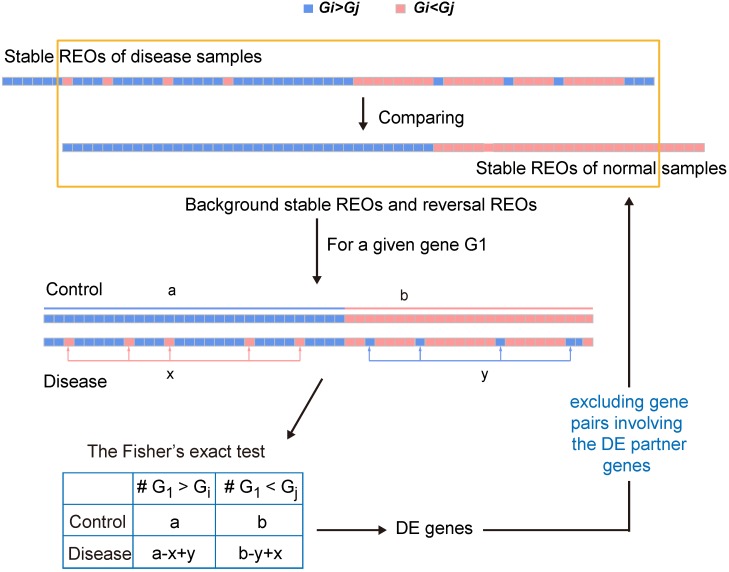
Figure 1.
A schematic diagram of the RankCompV2 algorithm. Focusing on the overlaps of the two lists of stable gene pairs, the stable REOs in the control samples are defined as the background stable REOs while the reversely stable REOs in the case samples are defined as the reversal REOs of the disease. For a given gene G1, a indicates the number of gene pairs with the REO pattern (G1 > Gi), and b indicates that with (G1 < Gj) in background stable REOs; x indicates the number of reversal gene pairs with shift (G1 < Gj→G1 > Gj) and y indicates the number of reversal gene pairs with shift (G1 > Gi→G1 < Gi) in reversal REOs of the disease 28, 30, 35.