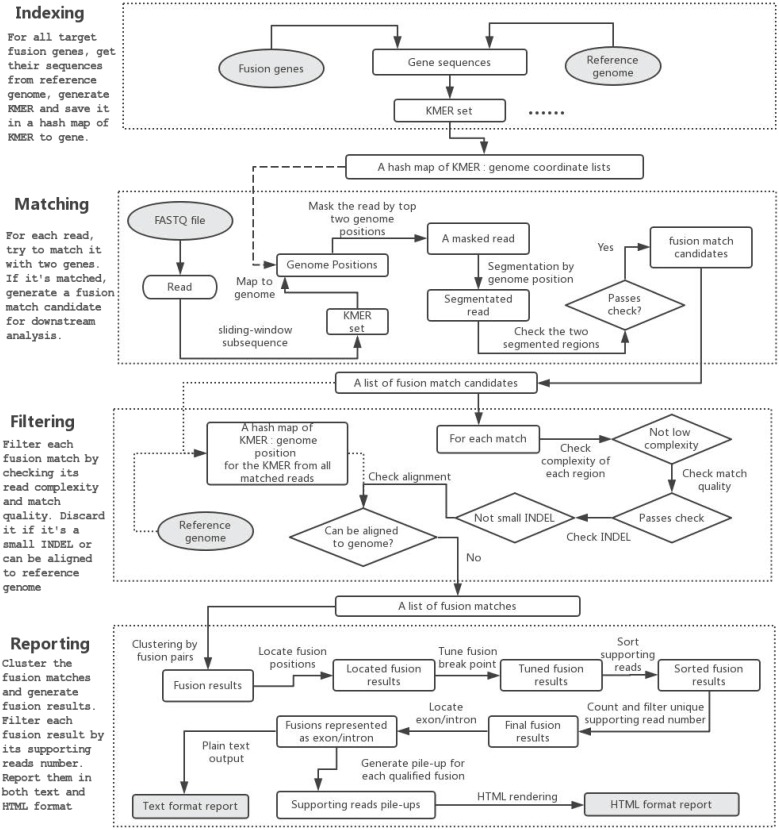
Fig 1.
The program flow of GeneFuse. Four steps are included in the workflow: indexing, matching, filtering, and reporting. In the indexing step, a hashmap of mapping to genes is computed. In the matching step, reads are mapped to the genes using the computed hashmap, and those that can be mapped to two genes are saved to fusion matches. In the filtering step, each fusion match is filtered by its read complexity, match quality, and other factors. Finally, in the reporting step, the detected fusions are validated, and the supporting reads for each fusion are piled up and rendered to an HTML page. The input and output files are then highlighted in grey.