Figure 7.
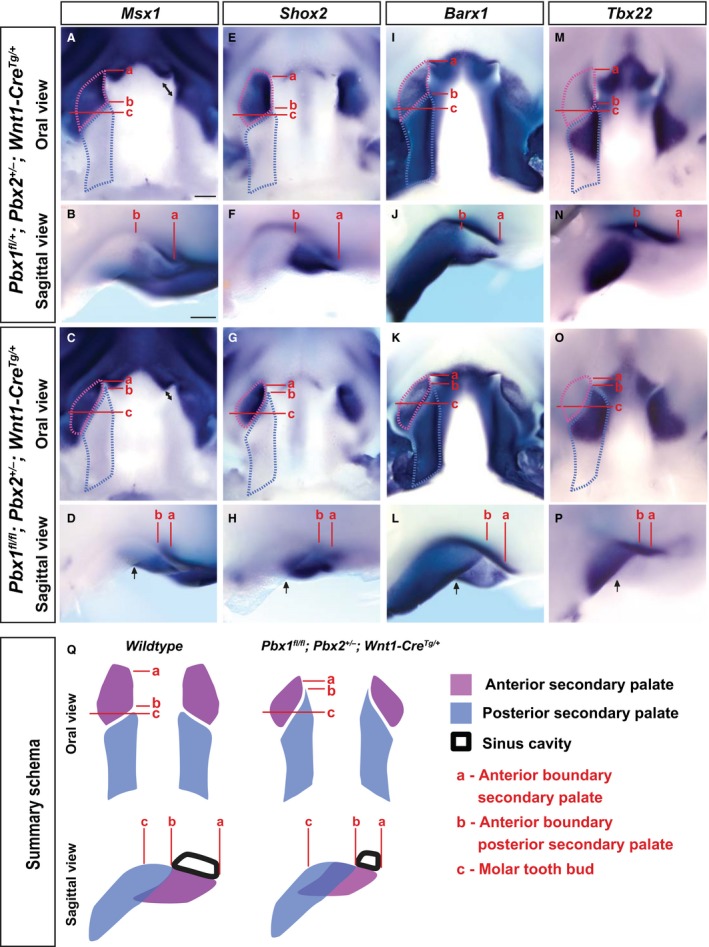
Altered positioning of transcriptional domains along the palatal shelf anterior‐posterior axis in Pbx CNCC mutant embryos. WISH reveals that spatial expression of anterior palate markers: Msx1 (A–D); Shox2 (E–H); epithelial‐Barx1 (I–L), as well as posterior palate markers: mesenchymal‐Barx1 (I–L) and Tbx22 (M–P), is perturbed in E12.5 CNCC mutants vs. controls (anterior is to the top in oral views and to the right in sagittal views). Anterior boundary of (a) anterior secondary palate, (b) posterior secondary palate and (c) molar tooth bud. All gene transcripts detectable at comparable levels in control (top) and mutant (middle) palates; however, spatial expression domains in anterior vs. posterior secondary palates show altered organization along A–P axis in mutants as compared with controls. Length of posterior secondary palate is similar between controls and mutants (light‐blue dashed outline), whereas the length of mutant anterior secondary palate is slightly reduced (light purple dashed outline; compare distance a–c). Significantly, rather than being positioned rostral to the posterior secondary palate as in controls (distance a–b > b–c), anterior secondary palate in mutants is more caudal (distance a–b < b–c) and lateral (black arrows) to posterior secondary palate. Maxillary dysmorphology accompanied by defective elongation of sinus cavity and nasal septum (black double‐headed arrows in oral view of Msx1 expression). Morphogenetic and anatomical defects in CNCC mutants summarized schematically (bottom row; Q). Consistent with highly coordinated outgrowth of the midfacial complex, positioning of anterior secondary palate (light purple) rostral to posterior secondary palate (light blue) is perturbed in mutants resulting in cleft palate and consequent reduced expansion of sinus cavity. Scale bar: 200 μm.
