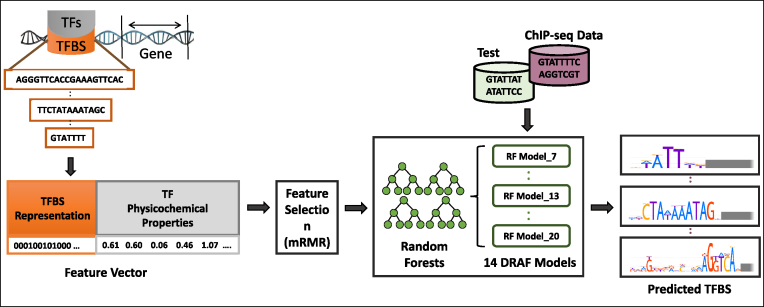
Figure 1.
The input data, training procedure and usage of the DRAF models for prediction of TF-TFBS links. Sequences of TFs and their TFBSs are represented in TF-TFBS links using physicochemical properties of TFs and binary representation of TFBSs. Then, the DRAF models were constructed for each group of TFs depending on the TFBS length. Finally, the DRAF models were tested using the holdout test dataset and another set of ChIP-seq peak data and their associated background datasets. The DRAF models aim at predicting which TF-TFBS link suggests a valid TFBS for a particular TF.