Fig. 1.
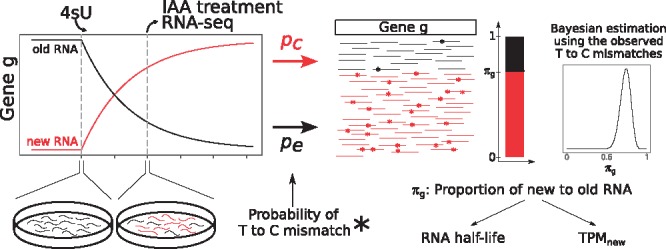
GRAND-SLAM overview. After a period of labeling with 4-thiouridine (4sU), RNA is extracted from cells, treated with IAA and sequenced. Shown is a theoretical timecourse of the abundances of new and old RNA for a gene g. IAA converts incorporated 4sU into cytosine analogs with an overall rate pc (including the incorporation rate, conversion rate and error rate), and uridines are sequenced as cytosine with an error rate pe. Based on the observed mismatches from T to C, the proportion of new to old RNA of gene g, πg, can be estimated using Bayesian inference. Estimates of πg can be transformed into estimates of the gene’s RNA half-life or relative abundance measures
