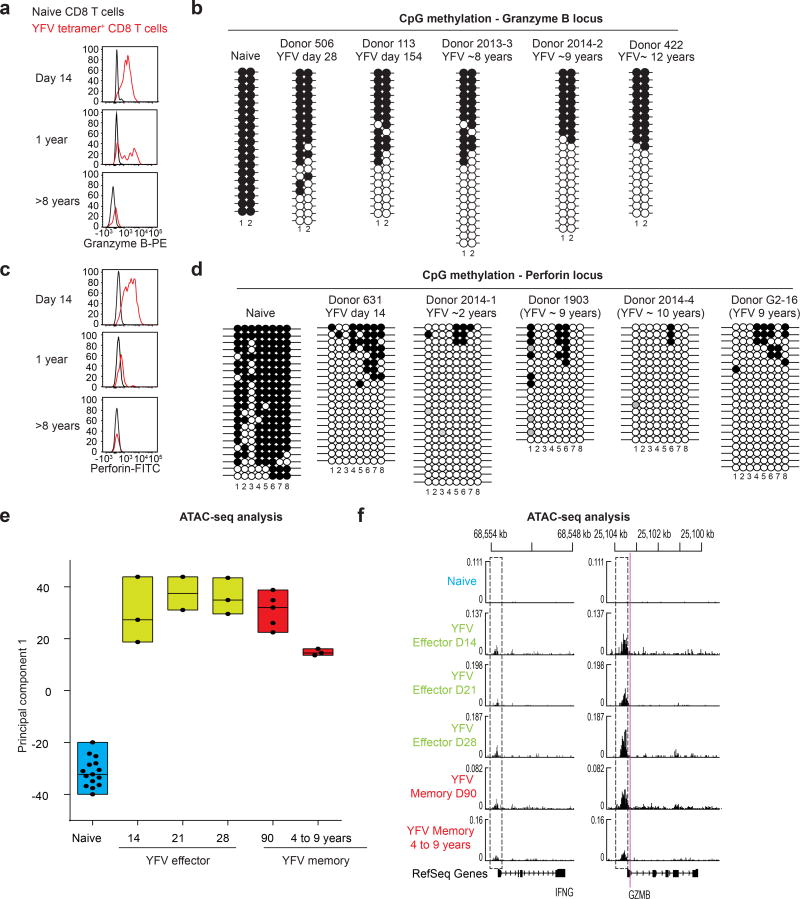
Figure 4. Long-lived YFV-specific memory CD8 T cells retain the epigenetic marks of their effector history.
a, c, Expression of granzyme B (a) and perforin (c) in naive and A2-NS4B214 effector, early memory and long-term memory CD8 T cells. PE, phycoerytherin; FITC, fluorescein isothiocyanate. b, d, Locus-specific bisulfite sequencing analysis of the CpG regions proximal to the promoter of GZMB (b) or PRF1 (d) in DNA from naive or tetramer+ CD8 T cells. Filled circles, methylated cytosine; open circles, non-methylated cytosine. e, ATAC–seq analysis of open chromatin in naive CD8 T cells (n = 15) and YFV-tetramer+ effector (n = 8), early memory (n = 5) and long-lived memory CD8 T cells (n = 3). Principal component analysis of ATAC–seq data summarizing the median, 25th and 75th percentiles for principal component 1. Each circle represents a sample from one donor. f, Traces of chromatin openness (y axis) near the IFNG and GZMB loci (x axis). The dashed rectangles represent open peaks that were significantly different (Wald test < 0.05) between at least two subpopulations. The purple line indicates the location of the CpGs investigated (in b) for methylation near the GZMB locus.