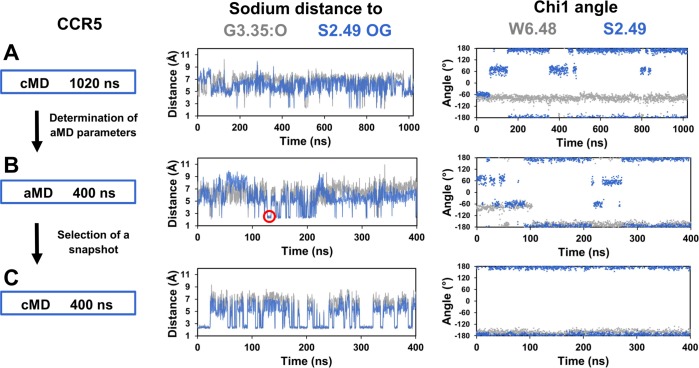
Fig 7. Alternative sodium binding site in CCR5.
In (A), classical MD simulations (cMD) of CCR5 was extended to 1.0 microsecond. Energy parameters from this trajectory were used to calculate acceleration parameters for the accelerated MD simulations (aMD) shown in (B). A snapshot with the sodium ion coordinated with the G3.35:O and S2.49:OG atoms (indicated by a red circle in the distance panel) was selected and used to initiate the classical MD simulations shown in (C). The left panel summarizes the protocol. The central panel shows the distances of the sodium ion to the G3.35:O and S2.49:OG atoms, colored in grey and blue, respectively. The right panel shows the chi1 dihedral angles of W6.48 and S2.49, colored in grey and blue, respectively.