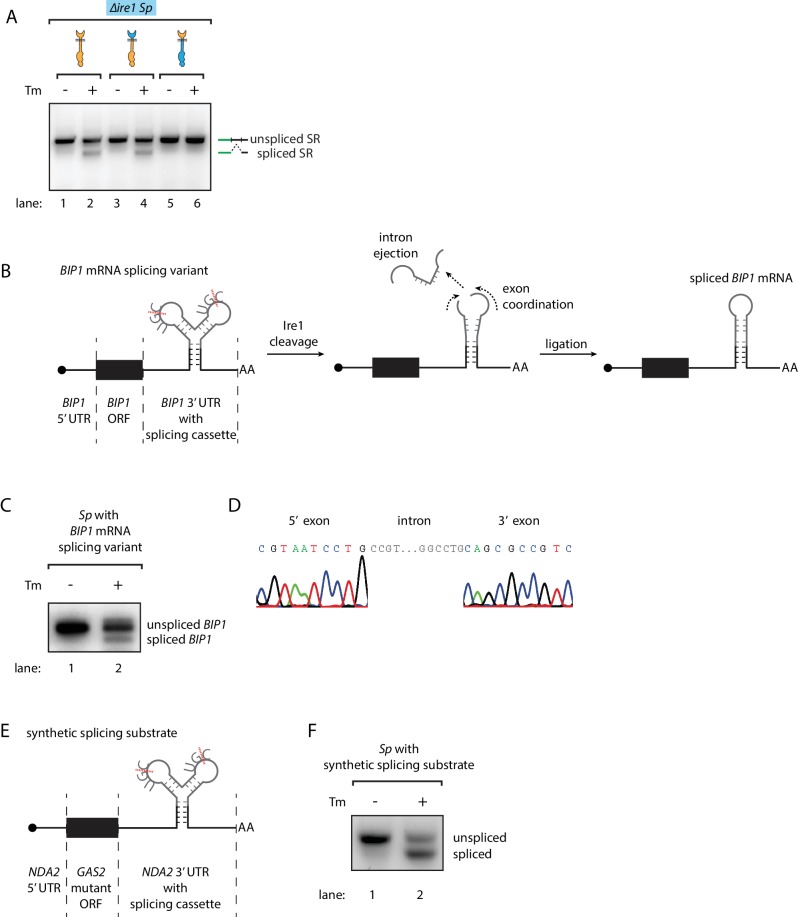
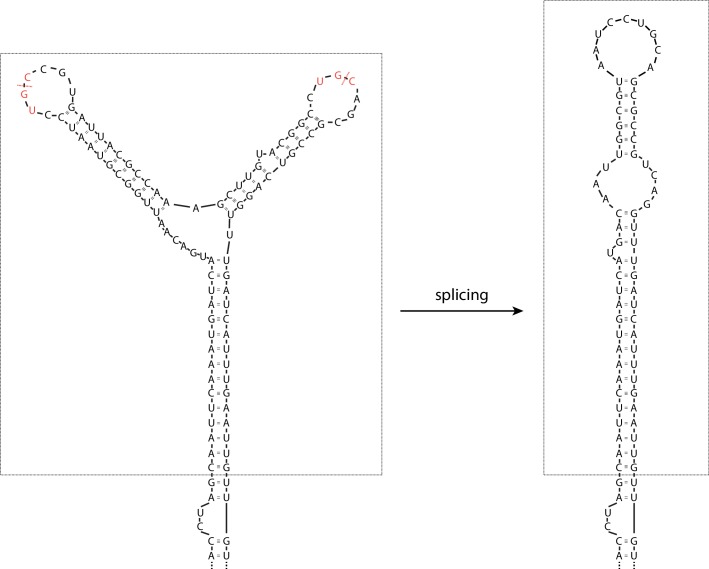
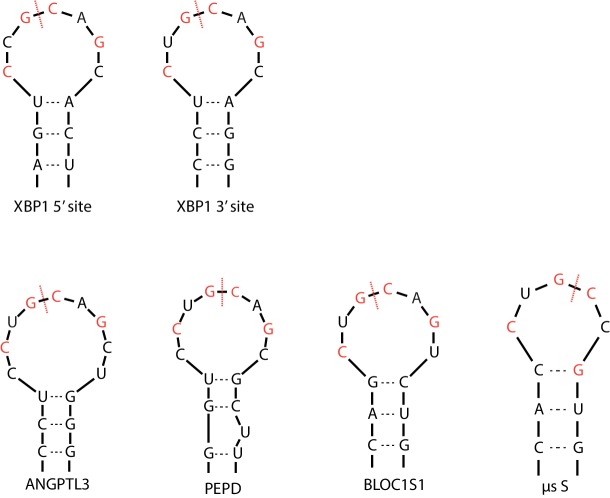
Figure 4. Engineering the Ire1-mediated non-conventional mRNA splicing in S. pombe cells.
(A) Measuring the non-conventional mRNA splicing in S. pombe cells, which were transformed with the S. cerevisiae HAC1 mRNA splicing reporter (SR) and the indicated Ire1 constructs. Cells were treated with 1 μg/ml Tm for 1 hr. (B) Illustration of the engineered S. pombe BIP1 mRNA splicing variant. (C) Measuring the non-conventional mRNA splicing of the engineered S. pombe BIP1 mRNA splicing variant. Experimental conditions are the same as those for Figure 4A. (D) Sequencing reads of the spliced BIP1 mRNA. The schematic illustration (E) and the splicing assay (F) of the synthetic splicing substrate in S. pombe. Cells were treated with 1 μg/ml Tm for 1 hr.