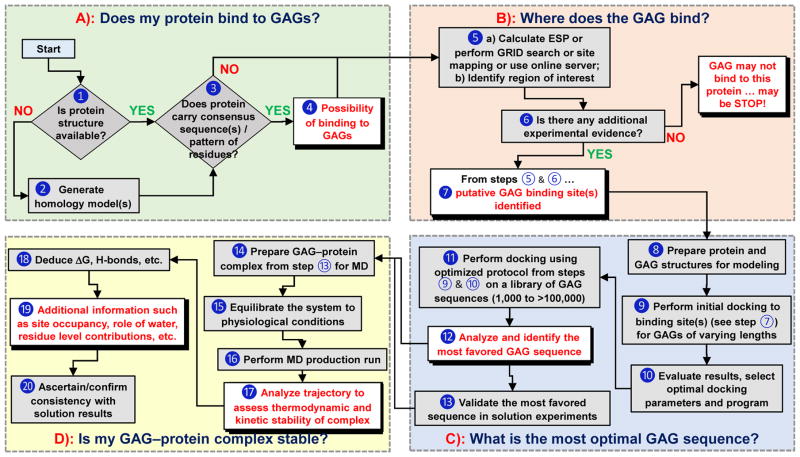
Figure 2.
A flowchart describing the use of computational approaches in addressing key questions on GAG–protein interactions (Panels A through D). Although shown in sequential format (A→B→C→D), it is not strictly necessary to rigorously follow this flowchart, especially if some information is already available for any of the steps. ➀ Sequence and atomic coordinates of a protein can be obtained from the protein data bank (www.rcsb.org). ➁ Homology model of a protein of unknown structure can be generated using programs such as Modeller (https://salilab.org/modeller/), Swiss-Model (https://swissmodel.expasy.org), etc. ➂ Consensus sequences include –XBBXBX-, -XBBBXXBX- (B = basic residue and X = hydropathic residue) [14], TXXBXXTBXXXTBB (T = turn),[41] CPC clif motif [43], clamp-like orientation of basic residues with beta sheet conformations [44]. ➃ If a protein satisfies step ➁, then it is likely to bind GAGs. ➄ a) Electrostatic potential (ESP) can be calculated using tools such as APBS from PyMol (https://www.pymol.org/), DeepView-Swiss-PdbViewer (http://spdbv.vital-it.ch/) and others. GRID search refers to protocol described by Goodford [50]. Site-mapping technique [30]. b) from step 5a we can identify basic site / subsite(s) ➅ Experimental evidence typically includes site directed mutagenesis, NMR, congenital mutation information, etc [52–54]. ➆ Putative GAG binding site(s) are identified based on results from ESP, GRID search, site-mapping techniques [17–20, 30]. ➇ This includes protonation, addition of hydrogens, modeling of missing residues and minimization of protein using a modeling software. GAG structures can be built using CHIMERA (https://www.cgl.ucsf.edu/chimera/) or GLYCAM (http://glycam.org/tools/molecular-dynamics/oligosaccharide-builder/build-glycan?id=8). ➈ Perform initial docking to site(s) of binding identified in step ➆ for various GAGs (HP, HS, CS, DS) of various lengths (dp2, dp4 and dp6) using either Autodock (http://autodock.scripps.edu), Autodock Vina (http://vina.scripps.edu), GOLD (https://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/), DOCK (http://dock.compbio.ucsf.edu/), MOE (https://www.chemcomp.com/MOE-Structure_Based_Design.htm), or other programs(refer supplementary information) ➉ Here GAG length, radius of site of binding, number of iterations, number of docking runs, type of docking program, etc. are evaluated and the best protocol is implemented in production run. ⑪ Perform repeated molecular docking using the optimized program and parameters from step ➉ for a library of GAG sequences. A library of GAG sequences can be obtained from the Desai lab (built using SPL scripts) [23,24]. Based on need, this library could have 1,000 to more than 100,000 unique sequences. ⑫ Analysis includes ranking of docked poses by calculating either RMSD, energy, score, non-bonded interactions, etc. and identify the most favored GAG equence(s). ⑬ Although typically not considered part of a computational program, validation of results in solution experiments obtained in ⑫ is extremely important. ⑭ Utilize the most favored GAG–protein complex from ⑬ and prepare initial coordinates for MD, which includes selecting force field, ensuring charge neutralization, immersing in an explicit box of solvent molecules, and minimizing the system. ⑮ Equilibration implies allowing the system to reach physiological conditions such as constant temperature and pressure (NPT/NVE) conditions. ⑯ This includes performing MD run for ~1 ns to ~1 ms, based on need, and collecting trajectories of data. ⑰ – ⑲ Analysis of trajectories may involve RMSD convergence, direct and water mediated H-bond interactions and their occupancies, binding free energy calculations (MMPBSA/MMGBSA), FEP, LIE and single residue energy decomposition calculations. ⑳ This involves ascertaining that computational deduction of thermodynamic stability on the basis of steps ⑰ through ⑲ is supported by some results in solution.