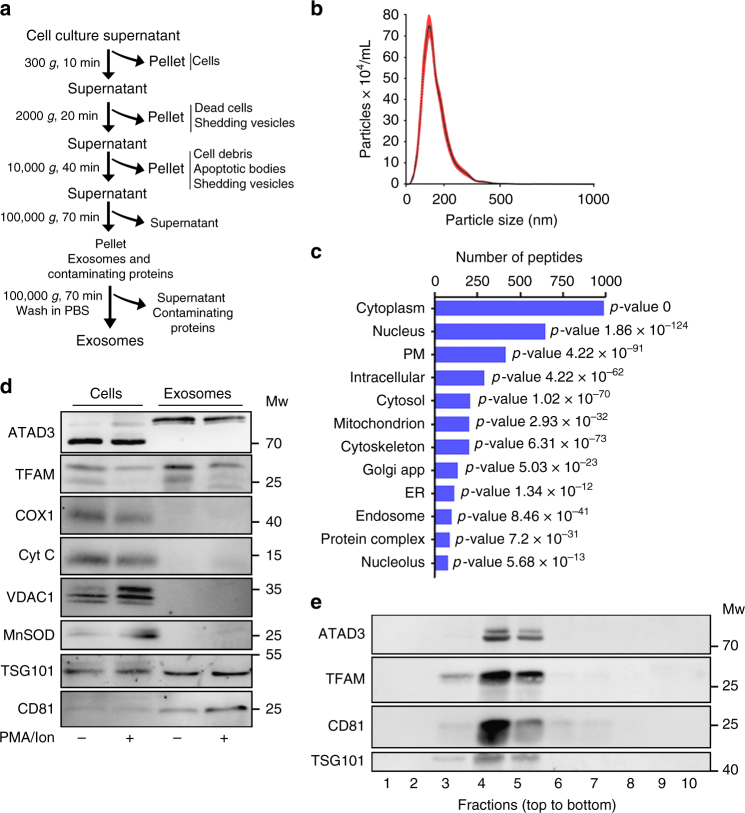
Fig. 1.
Exosomes shuttle mitochondrial proteins. a Differential ultracentrifugation protocol for isolating and purifying exosomes from cell culture supernatants. b Size distribution analysis by Nanoparticle Tracking Analysis (NTA) of purified EVs from primary human T lymphoblasts. c Gene ontology (GO) cellular component analysis of peptides identified in T cell EVs. d Western blot analysis of mitochondrial proteins in EVs obtained from the culture supernatant of primary human T lymphoblasts with or without treatment with phorbol myristate acetate (PMA) plus ionomycin. Cells and EVs were blotted for proteins associated with mtDNA (TFAM and ATAD3), for proteins located in the inner mitochondrial membrane (COX1 and Cytochrome C), the outer mitochondrial membrane (VDAC1), the mitochondrial matrix (mitochondrial manganese superoxide dismutase, MnSOD), and for the exosome markers TSG101 and CD81. e Western blot analysis of ATAD3, TFAM, and the exosome markers CD81 and TSG101 in sucrose fractions. EVs obtained from the culture supernatant of human T lymphoblasts were laid on a discontinuous sucrose gradient and floated by overnight centrifugation. Gradient fractions were collected and analyzed by immunoblot to reveal the distribution of mtDNA-binding proteins and exosomal proteins in the sucrose fractions from lower to higher sucrose density (left to right). Gels shown are representative out of three independent experiments