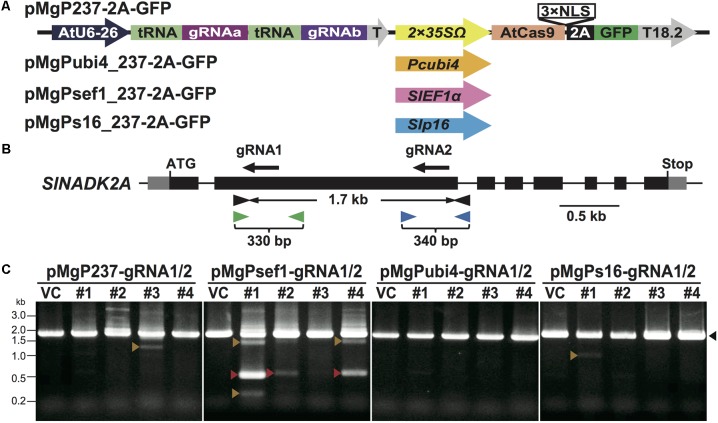
FIGURE 1.
Mutation analyses in the SlNADK2A gene of tomato calli using PCR and Cel-1 assay. (A) Schematic illustrating multiplex CRISPR/Cas9 vectors using different Cas9 promoters. AtU6-26: AtU6 snRNA-26 promoter, T: AtU6-26 terminator, 2 × 35SΩ: 2 × CaMV35S promoter with the omega enhancer sequence, Pcubi4: parsley Ubiqutin4-2 promoter, SlEF1α: SlEF1α promoter, Slp16: Slp16 promoter, AtCas9: Arabidopsis-codon optimized SpCas9, 2 A: 2 A self-cleavage peptide, NLS: nuclear localization signal, T18.2: Arabidopsis hsp18.2 terminator. (B) The gRNA target sites and primer sites in the SlNADK2A gene. The gRNA sites are shown as black arrows. Black, green, or blue arrowheads present primers used to amplify the 1.7 kb fragments including SlNADK2A-gRNA1 and SlNADK2A-gRNA2, the 330 bp fragments including SlNADK2A-gRNA1, or the 340 bp fragments including SlNADK2A-gRNA2, respectively. (C) Detection of deletion mutations between the target sites by PCR analysis. Black arrowheads; 1.7 kbp WT-sized bands, red arrowheads; deleted PCR fragments (0.5 kbp) of precisely the number of nucleotides between the two gRNAs, orange arrowheads; other size-deleted PCR fragments. Lane numbers indicate tomato callus lines.