Abstract
In this review, we introduce current developments in induced pluripotent stem cells (iPSCs), site-specific nuclease (SSN)-mediated genome editing tools, and the combined application of these two novel technologies in biomedical research and therapeutic trials. The sustainable pluripotent property of iPSCs in vitro not only provides unlimited cell sources for basic research but also benefits precision medicines for human diseases. In addition, rapidly evolving SSN tools efficiently tailor genetic manipulations for exploring gene functions and can be utilized to correct genetic defects of congenital diseases in the near future. Combining iPSC and SSN technologies will create new reliable human disease models with isogenic backgrounds in vitro and provide new solutions for cell replacement and precise therapies.
Keywords: induced pluripotent stem cells (iPSCs), site-specific nucleases (SSNs), zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated system 9 (Cas9)
Induced Pluripotent Stem Cell (iPSC) Technology
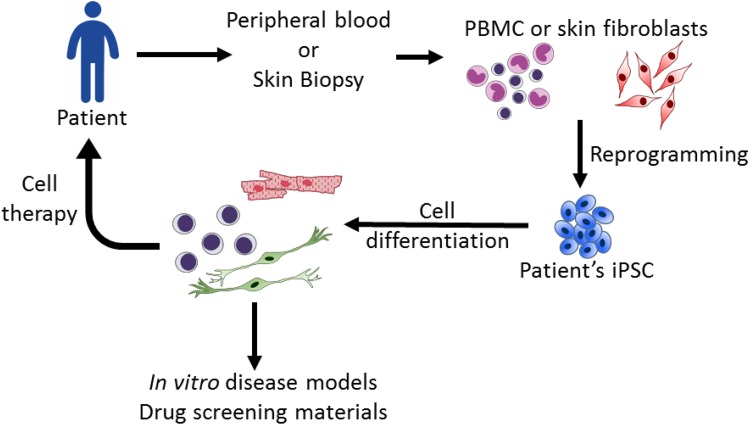
In 2006 and 2007, Dr. Takahashi and Dr. Yamanaka overexpressed four pluripotency-related transcriptional factors (octamer-binding transcription factor 4 (Oct4), Kruppel-like factor 4 (Klf4), sex-determining region y box 2 (Sox2), and c-myc) and successfully reversed mouse and human somatic cells back to a pluripotent status. These embryonic stem cell (ESC)-like cells are called induced pluripotent stem cells (iPSCs)1,2. iPSCs share similar properties with ESCs, including self-renewal, a normal karyotype, a 3-germlayer cell formation and germline transmission ability1,2. These unique advantages of ESC-like properties and personalized fabrication from somatic cells rapidly garnered world-wide attention to this technology. Accumulative research has steered the fundamental improvement of the efficacy of iPSC establishment, including culture conditions, optimal cell sources2,3, vector designs4–8, and reprogramming assistance by proteins and small molecules9–11. Notably, Dr. Hou reported the success of iPSC production by chemical induction without the introduction of Yamanaka factors12. Currently, iPSCs are widely applied in basic research and have become a reliable in vitro platform for developmental studies, disease modeling and drug screening (Fig. 1).
Fig. 1.
Applications of induced pluripotent stem cell (iPSC) technology. iPSCs derived from patients can be differentiated into specific cell lineages to recapitulate cytopathies for disease studies and potential drug screening. For therapies, iPSC-derived cells can provide materials for transplantation.
Genome modifications in pluripotent stem cells (PSCs) will fundamentally improve the feasibility for researchers to delineate the cell fate, patterning of gene expression, and niche environment regulation at different developmental stages or in 3D organoid architecture. The following text will briefly introduce the genetic editing tools through both random insertion and site-specific modification.
Development of Genome Editing Tools: Genome Modifications Before Site-Specific Nucleases (SSNs)
For genetic modification, there are two major strategies, random insertion and site-specific targeting. For random insertion, lentiviruses13 and retroviruses14 are the most commonly used vectors. Other well-known random insertion tools are transposons, including Sleeping Beauty15, piggyBac16, and others. Through the help of the transposase protein, DNA fragments surrounded with a terminal repeat sequence can be randomly inserted into a host genome. Different from lentiviruses or retroviruses, the transposon can be excised from the host genome via re-expression of transposase and reverse back to transgene-free cell clones15,16.
Foreign DNA fragments can be inserted into the host cell genome for different purposes, like gene-specific reporters and gene overexpression. Despite the convenience of the genetic tools, this approach has several shortcomings. First, the random inserted segments may induce mutagenesis in host cells. In addition, the expression level of random inserted genes may be different from the natural expression level of host cells. In some cases, the inserted genes may be silenced, depending on the insertion sites of chromosomes.
Compared with random insertion strategies, site-specific DNA targeting provides higher stability and accuracy for genetic research. For instance, transcription regulatory elements of most genes are still not clear and restrict the application of transgenic systems to genetic function research. Site-specific DNA targeting can overcome these defects of the transgenic approach and become powerful tools for genetic research and therapies. To implant a foreign DNA segment into a specific position of a chromosome, homologous recombination (HR)-based targeting is the traditional approach. Two homologous arms on the 5’ and 3’ ends of foreign DNA are essential for spontaneous HR17. Site-specific HR is widely used in mouse ESCs (mESCs) for generating knock-in/knockout mice18. Several genetically modified human PSC (hPSC) lines have also been established for disease models. These strategies have also been used to establish gene-specific reporter hPSCs, such as Oct4 (a pluripotent specific marker) and Oligo2 (a neuroglia specific marker), for cell differentiation research or specific cell lineage purification19,20.
Although the HR approach is widely applied to mESCs, this genetic targeting approach is limited in hPSCs. The major challenge is the dissociation-induced cell death of hPSCs. Most hPSCs undergo anoikis and die after cell dissociation due to loss of the cell–cell surface cadherin junction21,22. This property not only reduces the DNA transfection or electroporation efficiency, but also influences the efficacy to obtain targeted hPSC lines from a single cell. Recent evidence indicates that hPSC dissociation-induced cell death can be inhibited by adding Y27632, which blocks the activation of Rho/ROCK signaling pathway and sustains the cytoskeleton architecture21–23. In addition, this cell death can also be dramatically attenuated when the hPSCs status is converted to naïve state. Naïve hPSCs share similar features with mESCs, including cell morphologies, germline markers expression and high survival rates in single cell status24,25. These properties of naïve hPSCs may benefit the improvement of genome editing efficiency in hPSCs. Moreover, newly-developed feeder-free and defined hPSC culture medium fundamentally accelerate the generation of genetic modified hPSCs by avoiding the disturbance of feeder cells during gene transfer and increasing the homogeneity of expanded cultured cells26–28.
The Development of Genome Editing Tools: Site-Specific Nuclease-Mediated Genome Editing
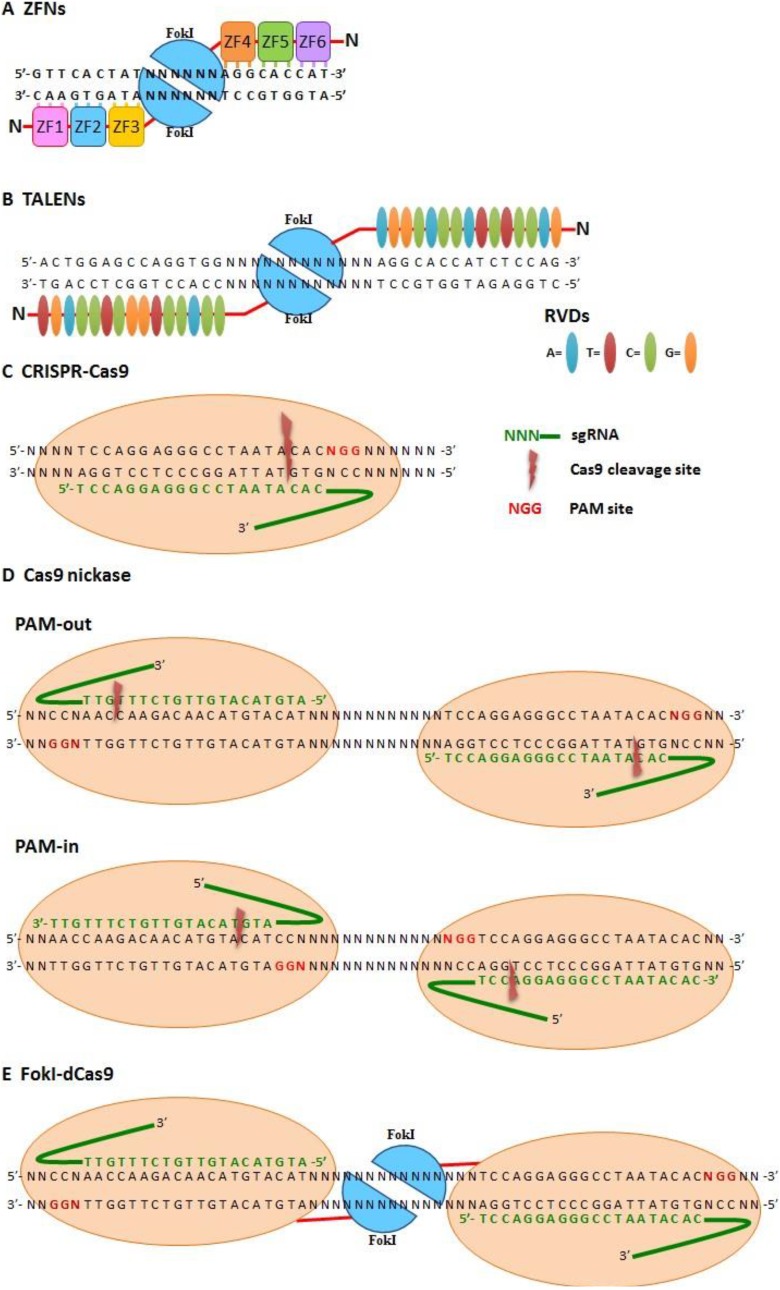
The efficacy of genetic editing by HR can be enhanced by creating DNA break by SSNs. SSNs can initiate the DNA repair system, including both homology-directed repair (HDR) and non-homologous end joining (NHEJ). The most widely used SSNs for genome editing are zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated system (Cas9)29 (Fig. 2).
Fig. 2.
Site-specific nuclease (SSN)-based genome editing tools. (A) Zinc-finger nucleases (ZFNs). Two zinc-finger nucleases (ZFNs) cooperate for site-specific recognition with a 3-bp pairing/zinc finger and dimerized FokI to create double-strand breaks (DSBs) on DNA. (B) Transcription activator-like effector-based nucleases (TALENs). Two TALENs cooperate for site-specific recognition with 1-bp pairing/repeat variable di-residues (RVDs) and dimerized FokI to create DSBs on DNA. (C–E) Cas9-based SSN system. sgRNA recognizes specific sites via Watson–Crick pairing and cleavage target DNA (C) with wild-type Cas9 (which makes DSB), (D) Cas9 nickase (double-strand nicking with PAM-out and PAM-in orientations), and (E) FokI-dCas9 (which creates DSBs with dimerized FokI).
The zinc finger domain of transcription factor, a DNA-binding motif that recognize and bind to specific DNA sequences, was artificially conjugated with DNA nuclease as zinc-finger nuclease (ZFN) for human genome targeting in 200530. ZFNs are composed of tandem repeating DNA-binding domain which recognize three bases of nuclear acids. For the balance of DNA specificity and targeting efficiency, 3 to 6 zinc fingers are the most suitable length for genome editing. The most common nuclease of ZFN is the FokI restriction enzyme31,32 (Fig. 2A). ZFNs are widely used for human genome editing33. A clinical trial using ZFNs to modify the chemokine receptor CCR5 for human immunodeficiency virus (HIV) therapy is now in progress29,34–37.
Although ZFN is a powerful tool for genome editing, there are still some limitations for this SSN. First, the optimal couple between the zinc finger domain and the nucleotides is a challenge. It is a tremendous work to tailor proper amino acid sequence of the DNA-binding domain for properly fitting the variables of 3-mer DNA sequence. Second, the pairing efficiency of nucleic acids and zinc fingers may be influenced by up- and downstream nucleic acid sequences and zinc finger compositions. Finally, the low binding specificity may also cause off-target mutations and targeting failure.
Transcription activator-like effector (TALE) nucleases (TALENs) is another common SSN for genetic targeting and contain both DNA-binding domain and nuclease domain. The DNA-binding domain of TALE proteins, derived from the plant bacterium, Xanthomonas, are composed of a series of repeating residues with 33∼35 amino acids. The 12th and 13th amino acids within a TALE residue are variable and play major roles in DNA recognition, called repeat variable di-residues (RVDs). Each RVD can specifically recognize a nucleic acid (Asn-Gly for thymine, His-Asp for cytosine, Asn-Ile for adenine, and Asn-Asn for guanine)38–40. Combinations of continuous designed TALE theoretically can recognize all DNA sequences. After DNA-binding, the FokI nuclease can break DNAs with DSB nicks and induce HDR or NHEJ DNA repair reactions41 (Fig. 2B).
Compared with ZFNs, TALENs have only four types of RVDs to cover the four nucleotides. This advantage makes it much easier to generate TALEN-targeting clones for gene targeting. To overcome difficulties of assembling continuously repeating residues, many strategies have been developed for TALEN assembly, including the Golden Gate method, Platinum Gate method, and ligation-independent cloning (LIC)42–46.
The Development of Genome Editing Tools: CRISPR/Cas9
CRISPR/Cas9 system, the most common SSN from Streptococcus pyogenes, is first discovered from a microbial adaptive immune system against phage infection47 (Table 2). There are three components of CRISPR/Cas9-mediated DNA recognition and cleavage: Cas nuclease, CRISPR (cr)RNA and trans-activating crRNA (tracrRNA)48–50.
Table 2.
Comparison of site-specific nuclease (SSN) genome editing tools of zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated system 9 (Cas9).
| SSN system | DNA recognition type | Targeting flexibility | Easy to design and use | Targeting efficiency | Off-targeting effects |
|---|---|---|---|---|---|
| ZFN | Protein guide, trimer | + | + | + | ++ |
| TALEN | Protein guide, monomer | +++ | ++ | ++ | +++ |
| CRISRR/Cas9 | RNA guide, monomer | +++ | +++ | +++ | + |
Cas9 contains two nuclease domains, HNH and RuvC, to make a DSB48–50. For CRISPR/Cas targeting, tracrRNA promotes crRNA maturation and conjugates with processed crRNA to form small-guide (sg)RNA for targeting site recognition. The specificity of the gene targeting depends on the pairing of targeting sequencing (also named as protospacer) with sgRNA and protospace-adjacent motif (PAM), which interact with Cas9 protein48–50 (Fig. 2C).
Accumulative evidence indicates that off-targeting effect of CRISPR/Cas9 mainly causes by the high tolerance in the sgRNA sequence (up to five mismatches) and the DSB-induced NHEJ repair. To decrease the off-targeting rate and increase the accuracy of genetic modification, Cas9 nickase is generated by inactivating the RuvC nuclease region of Cas9 (D10A mutant in RuvC) to trigger a DNA single-strand break and predominant repair by the high-fidelity base excision repair pathway. Paired Cas9 nickases with reverse orientation DNA-binding are able to make double-strand nicks (Fig. 2D). This modified Cas9 nickase approach can improve targeting specificity by more than 50–1500-fold compared with wild-type Cas9, and also lowers the off-targeting rate51–53. Truncated sgRNAs may also reduce off-targeting effects of CRISPR/Cas954.Moreover, replacing the Cas9 with FokI, called fCas9, increased targeting specificity more than 140-fold compared with the wild-type Cas9 in human cells (Fig. 2E)55,56.
Application of SSNs in Clinical Trials
For gene therapy, the development of efficient genome targeting tools provides possibilities for clinical applications. Corrected cells may be helpful for transplantation therapies. The first application of SSNs in clinical trials was to treat acquired immune deficiency syndrome (AIDS). For preclinical AIDS therapy, researchers applied genome editing tools to generate mutations in HIV co-receptor CCR5 that blocks HIV infection and proliferation on CD4 T-cells34–36,57. The modified T-cells are resistant to HIV infection. Based on the positive results, a phase I study which used ZFNs to modify CCR5 of patient CD4 T-cells for autologous transplantation was initiated in 2009. A total of 12 patients with chronic aviremic HIV infection were enrolled, and 6 of them were treated with CCR5-edited CD4 T cell autologous transplantation. Results showed that HIV RNA levels in one patient (this patient was heterozygous for CCR5 delta 32) became undetectable and decreased in most patients. This trial demonstrated the safety and curative efficacy of this strategy. Notably, replenished CCR5 edited T-cells are required for long-term therapy to overcome the short-term survival of edited T-cells or alternatively using hematopoietic stem cells as the next genetic engineered target for providing long-term T-cells.
Application of hPSCs in Clinical Trials
Clinical trials of PSCs have shown the safety and efficacy of the transplantation of PSC-derived differentiating cells, including the treating for age-related macular degeneration (AMD), type I diabetes, Parkinson’s disease (PD), and myocardial infarction (Table 1)58.
Table 1.
List of current human pluripotent stem cell (hPSC)-based clinical trials.
| Disease | Engrafted cell type | PSC type | hPSC source | Current clinical trial phase |
|---|---|---|---|---|
| Spinal cord injury | Oligodendrocyte precursor cell | ESC | Allogenic | I |
| Dry age-related macular degeneration & Stargardt disease | Retinal pigment epithelium | ESC | Allogenic | I/II |
| Wet age-related macular degeneration | Retinal pigment epithelium sheet | iPSC | Autologous/allogenic | I |
| Parkinson disease | Dopaminergic neuron | iPSC | Allogenic | I |
| Type I diabetes | β-cell progenitor | ESC | Allogenic | I/II |
| Myocardial infarction | Cardiomyocyte | ESC | Allogenic | I |
ESC: embryonic stem cell; hPSC: human pluripotent stem cell; iPSC: induced pluripotent stem cells; PSC: pluripotent stem cells.
The first clinical trial using human ESCs (hESCs) targeted spinal cord injury, supported by Geron company. This phase I clinical trial was initiated in October 2010. hESC-derived oligodendrocyte progenitor cells (GRNOPC1) were used to treat spinal cord injury patients in the subacute stage59. Unexpectedly, Geron halted this program in 2013. However, Asterias re-initiated this clinical trial using the same cell type in 2016 due to the success of using hESCs in AMD.
For type I diabetes mellitus (DM), a phase I/II clinical trial using hESC-derived pancreatic precursor cells60 was initiated in October 2014 by the company Viacyte. Type I DM is caused by the loss of β-islet cells, and patients need to inject insulin for the rest of their lives. In this trial, researchers used a biocompatible capsule to protect hESC-derived pancreatic precursor cells from immune rejection61. Animal studies revealed that these encapsulated precursor cells can sense the blood glucose level and secrete insulin over the long-term without the formation of a teratoma. However, detailed results from human clinical trials are still under investigation.
hESC-derived cardiomyocytes have potential for myocardial infarct therapy. In non-human primate preclinical studies, transplantation of ESC-derived cardiomyocytes improved heart function after a myocardial infarct with no ventricular arrhythmia- related problems62. For the phase I clinical trial, hESC-derived cardiac progenitor cells were merged with fibrin to form a patch, which was then engrafted onto the damaged area of the heart. After 3 months, the heart function had improved without arrhythmias being observed63,64.
The functional improvements of PSC-derived cells were first validated in the AMD studies. AMD, the most common type of blindness in the elderly, is caused by the progressive degeneration of the retinal pigment epithelium (RPE). Stargardt macular dystrophy (SMD), also caused by RPE degeneration, is a juvenile-onset inherent disease caused by mutations of ABCA4, ELOVL4, and PROM1. The phase I results in AMD and SMD patients showed that no tumorigenesis, immune rejection, and RPE outgrowth were observed after the engraftment of ESC-derived RPE sheets. Notably, long-term observation of these phase I/II trials revealed that transplanted RPE cells were integrated with the layer of photoreceptors and significantly recovered the vision spectrum in recipients65–67.
Clinical trials for AMD using iPSCs were carried out in Japan. The phase I trial began in Japan in September 2014, initiated by the Kyoto University’s Center for iPSC Research and Application, Center for Developmental Biology RIKEN (RIKEN CDB), and Kobe City Medical Center General Hospital. This was the world’s first clinical trial using patient’s own iPSCs for autologous transplantation68,69. Long-term observations revealed no tumorigenesis or immune rejection. In addition, transplantation of the derived RPE sheets ameliorated the vision deterioration in the patients. More clinical patients will be recruited for the safety evaluation of iPSC-based cell therapy.
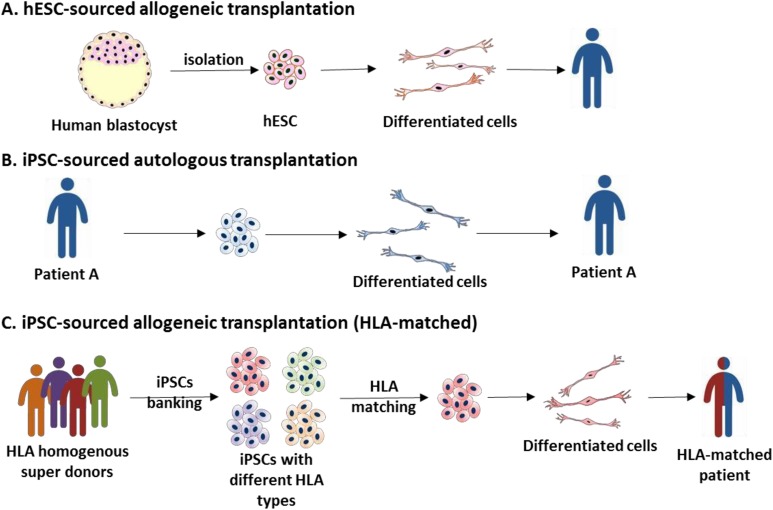
In the above-mentioned clinical trials, most applied hESCs were allogenic. Allogeneic transplantations can only be applied in some immune privilege sites, like the eyes and spinal cord. Some special strategies need to be considered to avoid immune rejection61. Personalized iPSC lines can resolve immune-related problems. However, generating and characterizing personalized clinical-grade iPSC lines are expensive and time consuming. Allogenic transplantation with human leukocyte antigen (HLA)-matching donors is a standard procedure of organ and bone marrow transplantation. It is an ideal strategy to establish and bank a few HLA-homozygous super-donor iPSC lines for clinical applications without the drawback of severe immune rejection (Fig. 3).
Fig. 3.
The source of human pluripotent stem cells (hPSCs) for clinical therapies. (A) Currently, most clinical trials use allogeneic human embryonic stem cells (hESCs) for transplantation without human leukocyte antigen (HLA) matching. (B, C) In Japan, two clinical trials used autologous and HLA matching allogeneic-induced pluripotent stem cells (iPSCs) as cell sources. For burden consideration, HLA homogenous iPSC banking is the optimal cell supplement source.
Genome Editing Tools Expand the Application Horizon of iPSC-Based Research and Disease Modeling
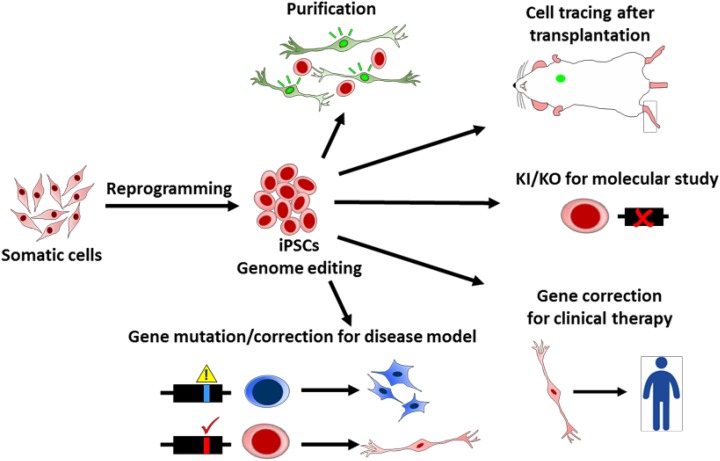
PSCs provide an in vitro model to recapitulate human development processes and cell lineage differentiation, especially for the early embryo stage. With genome modifications, researchers can delineate the cell fate, gene expression timing, and niche environment regulation at different developmental stages and 3D organoid architecture. Differentiating PSCs also can serve as a platform to address single or multiple gene functions and their physiological roles in vitro by gene mutations, deletions, or replacement 19,20,50,70–81 (Fig. 4).
Fig. 4.
Applications of genome editing tools for induced pluripotent stem cell (iPSC) technology. Genetic technologies provide kinds of application potentials for iPSC research. For example, genome editing tools are used to generate reporter cells for cell purification and tracing, knock-in and knockout (KO) for molecular studies, mutations or corrections for disease modeling, and clinical therapies.
Genome editing tools also provide the feasibility to generate isogenic iPSCs for accurate disease modeling. Specific mutations of disease iPSCs can be artificially fabricated or corrected by genome editing tools to generate isogenic disease iPSCs without the interference of cell resources, random mutations and epigenetic variation during the iPSC establishment. This genotype/phenotype validation by SSN-mediated gene correction or introduction consolidates the roles of candidate genetic loci in patients-derived iPSCs.
SSN-mediated genetic corrections of patient-derived PSCs are applied to Parkinson’s disease82, Niemann–Pick type C (NPC) disease83, sickle cell disease84,85, β-thalassemia86–88, Rett syndrome89, cystic fibrosis90–92, and α1-antitrypsin deficiency93. CRISPR/Cas9 was also used to correct mutations of MYO15A in hair cell-like cell deficiency94, chromosome 7q deletion in myelodysplastic syndromes95, and COL7A1 in recessive dystrophic epidermolysis bullosa (RDEB)96. For Down syndrome trisomy 21 correction, an inducible XIST transgene was introduced into the DYRK1A locus of chromosome 21 with a ZFN-editing tool. This modification successfully silences the chromosome 21 and forms the chromosome 21 Barr-body to maintain genetic expression balances97. Combining of these two technologies also provide us to generate gene corrected iPSCs of large-scale chromosome abnormal diseases like Duchenne muscular dystrophy (DMD)98,99. The above examples demonstrate that SSN- mediated genetically corrected hPSCs not only provide research materials but also potentially serve as healthy cell sources for autologous grafting.
Future Direction of SSN/hPSC-Combined Applications
Integrating both fast-evolved genome editing tools and standardized hPSCs fundamentally accelerates the scientific progress on the human developmental studies, disease modeling, specific cell tracing/isolation and clinical cell therapy19,20,50,70–81. The exogenous factors-directed differentiation from PSCs basically follows the developmental principles and reflects early embryonic formation, enabling the hPSC as an ideal model to investigate early human development. Given the high efficacy of CRISPR/Cas9-mediated on single gene knockout, the impact of a disrupted gene can be traced in the hPSCs and their derivatives. Investigating the mutant effects in spherical 3D organoid culture of PSCs can further broaden the affected spectrum of the cell–cell interaction, such as cell migration, niche environment and tissue organization100–103. By the stable expression of inducible Cas9 at the AAVS1 locus, similar to the ROSA26 locus in mice, precisely inactivating the gene expression at desired developmental stages can also be achieved104. This constitutive expression of Cas9 endonuclease (iCRISPR) significantly improves the efficacy of multiple gene inactivation and facilitate the exploration of orthologous genes simultaneously in developmental studies. Moreover, the CRISPR/Cas9 system can be further modified to be a gene repressor or converted to be a gene activator module to delineate the regulatory complex and gene function in specific cell types105,106. These highly evolved and versatile genetic editing tools will substantially shape the hPSCs as a new platform for early human developmental study.
Accumulative evidence indicates that familial genetic mutations in the patient-derived iPSCs are high penetrable and can faithfully recapitulate the disease phenotype in the derivative target cells from iPSCs100,107–122. The disease cytopathology can be corrected by the TALEN or CRISPR/Cas9-mediated HR123–127. Given the power of CRISPR/Cas9 system, researchers can modify the disease progress and severity by introducing gene deletion or repair using synthetic oligonucleotides in isogenic iPSCs. The interaction network of risk factors can also be delineated by multiplex gene knockout. The combined CRISPR/Cas9 and PSCs also provide a valuable system to investigate the accumulation effects of common but low-risk genetic factors for the progress of non-familial and idiopathic diseases, especially for the neurodegeneration, diabetes, atherosclerosis and cancers. Recently, whole-genome gRNA libraries (genome-wide CRISPR knockout screen [GeCKO]) have been established to dissect gene function in several cancer lines and hPSCs128,129. These innovative approaches will continuously provide fundamental and comprehensive understanding for the etiology of multiple-hit idiopathic diseases, paving the way for an optimized treatment strategy for patients.
Fabricating PSCs with genetic editing tools can extend the application of precision medicine by introducing specific tag on the cells and correcting gene mutation in PSC-derived cells. Expressing single or multiple specific reporter proteins under cell specific promoters can assist the cell tracing and cell sorting for further cell analysis and transplantation19,20,70–75. Currently the clinically applied PSC-derived RPE sheet for AMD is manually isolated without cell sorting. Genetic labeling of PSC-derived cells may be required in the future for enriching desired cell population, such as dopaminergic neuron precursors and hematopoietic stem cells, to facilitate the tracing of cell fate, cell survival rates and tumorigenesis in vivo.
The advantage of ease, cost, efficacy and versatility warrants CRISPR/Cas9 system as a suitable tool for human gene therapy. Several clinical trials based on genome editing and PSC technologies also reveal the possibilities to bring SSN/PSC-combined transplantation therapies to the real world. Not only for genetic disease, SSN/PSC-combined technologies also have potential for HIV and cancer therapies130. However, to apply PSCs and genome editing tools for clinical use, there are still some serious challenges on the road ahead that need to be overcome.
Large-scale production of clinical grade, high-purity engineered specific cell lineages is the first challenge for clinical applications. iPSC generation, specific cell lineages differentiation and genome editing are effort and time consuming, also a high cost processes. Especially in current Good Manufacture Practices (cGMP) norm. To streamline the procedure of SSN/PSC-based therapies, cGMP grade HLA iPSC banking is an imperative approach for the next decade. For clinical hPSC preparation, a bioreactor capable of large-scale amplification of high quality hPSCs is needed. Moreover, an efficient differentiation protocol to generate specific cell lineages is another big challenge131. For this issue, fluorescence-based cell sorting is an ideal option to purify specific cell lineages. For example, the surface marker, Corin and leucine rich repeats and transmembrane domains 1 (LRTM1) were used for dopaminergic neuron precursor purification132,133. As previously descripted, SSN-mediated cell lineage specific reporter can also provide an ideal solution for cell purification.
For clinical therapy, the most concerned issue is safety. The innate tumorigenic property of hPSCs is still a challenge. For clinical applications, how to identify low-tumorigenesis-risk iPSC clones and eliminate undifferentiated iPSCs before transplantation still has room to improve. Besides, the off-target risk of CRISPR/Cas9 is another challenge of safety. The high tolerance of mismatches for target recognition and strong DSB activity can create considerable off-targeting mutation, which dramatically hinders the clinical application on gene therapy. The researches using CRISPR/Cas9 for primary human CD4+ T-cells showed promising CCR5 specific targeting, negligible off-target mutation and HIV resistance134,135. However, testing CRISPR/Cas9 for homologous recombination-directed repair in tripronuclear embryos observed high unintended mutations in the genome of zygotes136. Although several approaches have developed to reduce the off-targeting drawbacks of CRISPR/Cas9 system, such as aforementioned Cas9-nickase, dCas9-FokI dimers and truncated sgRNA oligomer54,56, the clinical-grade gene-editing tools with high fidelity and low undesired mutations have to be developed and validated before the clinical applications in embryo or in PSC-based cell transplantation.
Recently, the discover of naïve-state hPSCs may benefit the differentiation of primordial germ cells (PGCs)137. Researchers also applied CRISPR/Cas9 to edit human embryo’s genome136. However, as technology advances, some ethical debates will arise. The SSN/PSC-based technologies may not only allow us to apply to transplantation therapies, but also allow us to generate modified germ cells. It is an important ethical concern that needed to be detailed discussed before applying these techniques for human germline modification and affect our next generation.
Conclusion
In conclusion, combining iPSCs and SSN genome targeting tools provides new insights for disease modeling and transplantation therapies. The tumorigenic and off-targeting issues of SSN/PSC-based therapies may be overcome in a near future and SSN/PSC-based therapies may be routinely clinical therapies. The future road map is challenging but hopeful.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Bio-innovation Center and financially supported by Buddhist Tzu Chi General Hospital (grant no. TCRD105-16).
References
- 1. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. [DOI] [PubMed] [Google Scholar]
- 3. Umegaki-Arao N, Pasmooij AM, Itoh M, Cerise JE, Guo Z, Levy B, Gostynski A, Rothman LR, Jonkman MF, Christiano AM. Induced pluripotent stem cells from human revertant keratinocytes for the treatment of epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra164. [DOI] [PubMed] [Google Scholar]
- 4. Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. Piggybac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, Yusa K, Bradley A, Meyers DJ, Mukherjee C, Cole PA, Cheng L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Z, Wu WS. Sodium butyrate promotes generation of human induced pluripotent stem cells through induction of the mir302/367 cluster. Stem Cells Dev. 2013;2(16):2268–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341(6146):651–654. [DOI] [PubMed] [Google Scholar]
- 13. Ikawa M, Tanaka N, Kao WW, Verma IM. Generation of transgenic mice using lentiviral vectors: A novel preclinical assessment of lentiviral vectors for gene therapy. Mol Ther. 2003;8(4):666–673. [DOI] [PubMed] [Google Scholar]
- 14. Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: Past, present and future. Oncogene. 2005;24(52):7656–7672. [DOI] [PubMed] [Google Scholar]
- 15. Aronovich EL, McIvor RS, Hackett PB. The sleeping beauty transposon system: A non-viral vector for gene therapy. Hum Mol Genet. 2011;20(R1):R14–R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson MH, Coates CJ, George AL., Jr Piggybac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15(1):139–145. [DOI] [PubMed] [Google Scholar]
- 17. Hasty P, Rivera-Perez J, Bradley A. The length of homology required for gene targeting in embryonic stem cells. Mol Cell Biol. 1991;11(11):5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hall B, Limaye A, Kulkarni AB. Overview: Generation of gene knockout mice. Curr Protoc Cell Biol. 2009;Chapter 19: Unit 19.12.1–19.12.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21(3):319–321. [DOI] [PubMed] [Google Scholar]
- 20. Xue H, Wu S, Papadeas ST, Spusta S, Swistowska AM, MacArthur CC, Mattson MP, Maragakis NJ, Capecchi MR, Rao MS, Zeng X, Liu Y. A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells. Stem Cells. 2009;27(8):1836–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, Sasai Y. A rock inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25(6):681–686. [DOI] [PubMed] [Google Scholar]
- 22. Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, Ueno M, Ishizaki T, Suemori H, Narumiya S, Niwa H, Sasai Y. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7(2):225–239. [DOI] [PubMed] [Google Scholar]
- 23. Chen G, Hou Z, Gulbranson DR, Thomson JA. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7(2):240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z, Mukamel Z, Krupalnik V, Zerbib M, Geula S, Caspi I, Schneir D, Shwartz T, Gilad S, Amann-Zalcenstein D, Benjamin S, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504(7479):282–286. [DOI] [PubMed] [Google Scholar]
- 25. Theunissen TW, Friedli M, He Y, Planet E, O’Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, Duc J, Cohen MA, Wert KJ, Castanon R, Zhang Z, Nery Huang Y, Jr, Drotar J, Lungjangwa T, Trono D, Ecker JR, Jaenisch R. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19(4):502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz- Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ludwig T, A Thomson J. Defined, feeder-independent medium for human embryonic stem cell culture. Curr Protoc Stem Cell Biol. 2007;Chapter 1: Unit 1C.2.1–1C.2.16. [DOI] [PubMed] [Google Scholar]
- 28. Nakagawa M, Taniguchi Y, Senda S, Takizawa N, Ichisaka T, Asano K, Morizane A, Doi D, Takahashi J, Nishizawa M, Yoshida Y, Toyoda T, Osafune K, Sekiguchi K, Yamanaka S. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep. 2014;4:3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012;9(8):805–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. [DOI] [PubMed] [Google Scholar]
- 31. Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to fok i cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. [DOI] [PubMed] [Google Scholar]
- 33. DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, Sancak Y, Cui X, Steine EJ, Miller JC, Tam P, Bartsevich VV, Meng X, Rupniewski I, Gopalan SM, Sun HC, Pitz KJ, Rock JM, Zhang L, Davis GD, Rebar EJ, Cheeseman IM, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20(8):1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. Gene editing of ccr5 in autologous CD4 T cells of persons infected with HIV. N Engl J 5 Med. 2014;370(10):901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28(8):839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo Riso P, Damo M, Pello OM, Holmes MC, Gregory PD, Gritti A, Broccoli V, Bonini C, Naldini L. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8(10):861–869. [DOI] [PubMed] [Google Scholar]
- 38. Bogdanove AJ, Voytas DF. Tal effectors: Customizable proteins for DNA 21 targeting. Science. 2011;333(6051):1843–1846. [DOI] [PubMed] [Google Scholar]
- 39. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of tal-type iii 24 effectors. Science. 2009;326(5959):1509–1512. [DOI] [PubMed] [Google Scholar]
- 40. Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by tal 26 effectors. Science. 2009;326(5959):1501. [DOI] [PubMed] [Google Scholar]
- 41. Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. [DOI] [PubMed] [Google Scholar]
- 42. Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7(1):171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. Flash assembly of TALENS for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom 1 TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cermak T, Starker CG, Voytas DF. Efficient design and assembly of custom TALENS using the Golden Gate platform. Methods Mol Biol. 2015;1239:133–159. [DOI] [PubMed] [Google Scholar]
- 46. Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation- independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31(1):76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18(5):573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. [DOI] [PubMed] [Google Scholar]
- 49. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas 14 systems. Science. 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;17 339(6121):823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off- target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to foki nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided foki nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, Chang JC, Bao G, Muench MO, Yu J, Levy JA, Kan YW. Seamless modification of wild-type induced pluripotent stem cells to the natural ccr5delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111(26):9591–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trounson A, DeWitt ND. Pluripotent stem cells progressing to the clinic. Nat Rev Mol Cell Biol. 2016;17(3):194–200. [DOI] [PubMed] [Google Scholar]
- 59. Chapman AR, Scala CC. Evaluating the first-in-human clinical trial of a human embryonic stem cell-based therapy. Kennedy Inst Ethics J. 2012;22(3):243–261. [DOI] [PubMed] [Google Scholar]
- 60. Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D’Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26(4):443–452. [DOI] [PubMed] [Google Scholar]
- 61. Lee SH, Hao E, Savinov AY, Geron I, Strongin AY, Itkin-Ansari P. Human beta-cell precursors mature into functional insulin-producing cells in an immunoisolation device: Implications for diabetes cell therapies. Transplantation. 2009;87(7):983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510(7504):273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Menasche P, Vanneaux V, Fabreguettes JR, Bel A, Tosca L, Garcia S, Bellamy V, Farouz Y, Pouly J, Damour O, Perier MC, Desnos M, Hagege A, Agbulut O, Bruneval P, Tachdjian G, Trouvin JH, Larghero J. Towards a clinical use of human embryonic stem cell-derived cardiac progenitors: A translational experience. Eur Heart J. 2015;36(12):743–750. [DOI] [PubMed] [Google Scholar]
- 64. Menasche P, Vanneaux V, Hagege A, Bel A, Cholley B, Cacciapuoti I, Parouchev A, Benhamouda N, Tachdjian G, Tosca L, Trouvin JH, Fabreguettes JR, Bellamy V, Guillemain R, Suberbielle Boissel C, Tartour E, Desnos M, Larghero J. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: First clinical case report. Eur Heart J. 2015;36(30):2011–2017. [DOI] [PubMed] [Google Scholar]
- 65. Schwartz SD, Hubschman JP, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R. Embryonic stem cell trials for macular degeneration: A preliminary report. Lancet. 2012;379(9817):713–720. [DOI] [PubMed] [Google Scholar]
- 66. Atala A. Human stem cell-derived retinal cells for macular diseases. Lancet. 2015;385(9967):487–488. [DOI] [PubMed] [Google Scholar]
- 67. Schwartz SD, Regillo CD, Lam BL, Eliott D, Rosenfeld PJ, Gregori NZ, Hubschman JP, Davis JL, Heilwell G, Spirn M, Maguire J, Gay R, Bateman J, Ostrick RM, Morris D, Vincent M, Anglade E, Del Priore LV, Lanza R. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: Follow up of two open-label phase 1/2 studies. Lancet. 2015;385(9967):509–516. [DOI] [PubMed] [Google Scholar]
- 68. Cyranoski D. Stem cells cruise to clinic. Nature. 2013;494(7438):413. [DOI] [PubMed] [Google Scholar]
- 69. 365 days: Nature’s 10. Nature. 2014;516(7531):311–319. Available at: https://www.nature.com/polopoly_fs/1.16562!/menu/main/topColumns/topLeftColumn/pdf/516311a.pdf. [DOI] [PubMed] [Google Scholar]
- 70. Bhatia S, Pilquil C, Roth-Albin I, Draper JS. Demarcation of stable subpopulations within the pluripotent hESC compartment. PLoS One. 2013;8(2):e57276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG. Targeting a GFP reporter gene to the mixl1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111(4):1876–1884. [DOI] [PubMed] [Google Scholar]
- 72. Forster R, Chiba K, Schaeffer L, Regalado SG, Lai CS, Gao Q, Kiani S, Farin HF, Clevers H, Cost GJ, Chan A, Rebar EJ, Urnov FD, Gregory PD, Pachter L, Jaenisch R, Hockemeyer D. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Reports. 2014;2(6):838–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mae S, Shono A, Shiota F, Yasuno T, Kajiwara M, Gotoda-Nishimura N, Arai S, Sato-Otubo A, Toyoda T, Takahashi K, Nakayama N, Cowan CA, Aoi T, Ogawa S, McMahon AP, Yamanaka S, Osafune K. Monitoring and robust induction of nephrogenic intermediate mesoderm from human pluripotent stem cells. Nat Commun. 2013;4:1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ruby KM, Zheng B. Gene targeting in a hues line of human embryonic stem cells via electroporation. Stem Cells. 2009;27(7):1496–1506. [DOI] [PubMed] [Google Scholar]
- 75. Goulburn AL, Alden D, Davis RP, Micallef SJ, Ng ES, Yu QC, Lim SM, Soh CL, Elliott DA, Hatzistavrou T, Bourke J, Watmuff B, Lang RJ, Haynes JM, Pouton CW, Giudice A, Trounson AO, Anderson SA, Stanley EG, Elefanty AG. A targeted NKX2.1 human embryonic stem cell reporter line enables identification of human basal forebrain derivatives. Stem Cells. 2011;29(3):462–473. [DOI] [PubMed] [Google Scholar]
- 76. Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27(9):851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5(1):97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–1306. [DOI] [PubMed] [Google Scholar]
- 80. Blair JD, Bateup HS, Hockemeyer DF. Establishment of genome-edited human pluripotent stem cell lines: From targeting to isolation. J Vis Exp. 2016;11(108):e53583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110(39):15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, Mungenast AE, Muffat J, Mitalipova M, Pluth MD, Jui NT, Schule B, Lippard SJ, Tsai LH, Krainc D, Buchwald SL, Jaenisch R, Lindquist S. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342(6161):983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, Gao Q, Mitalipova M, Jaenisch R. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann–Pick type c patient-specific iPS cells. Stem Cell Reports. 2014;2(6):866–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle 26 cell disease. Blood. 2011;118(17):4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LF, Artandi SE, Wernig M, Joung JK. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29(11):1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and Piggybac. Genome Res. 2014;24(9):1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ma N, Liao B, Zhang H, Wang L, Shan Y, Xue Y, Huang K, Chen S, Zhou X, Chen Y, Pei D, Pan G. Transcription activator-like effector nuclease (TALEN)- mediated gene correction in integration-free beta-thalassemia induced pluripotent stem cells. J Biol Chem. 2013;288(48):34671–34679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang Y, Zheng CG, Jiang Y, Zhang J, Chen J, Yao C, Zhao Q, Liu S, Chen K, Du J, Yang Z, Gao S. Genetic correction of beta-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 2012;22(4):637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J, Kwok SM, Feldman DA, Bateup HS, Gao Q, Hockemeyer D, Mitalipova M, Lewis CA, Vander Heiden MG, Sur M, Young RA, Jaenisch R. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13(4):446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Suzuki S, Sargent RG, Illek B, Fischer H, Esmaeili-Shandiz A, Yezzi MJ, Lee A, Yang Y, Kim S, Renz P, Qi Z, Yu J, Muench MO, Beyer AI, Guimaraes AO, Ye L, Chang J, Fine EJ, Cradick TJ, Bao G, Rahdar M, Porteus MH, et al. TALENS facilitate single-step seamless SDF correction of f508del CFTR in airway epithelial submucosal gland cell-derived cf-iPSCs. Mol Ther Nucleic Acids. 2016;5:e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Crane AM, Kramer P, Bui JH, Chung WJ, Li XS, Gonzalez-Garay ML, Hawkins F, Liao W, Mora D, Choi S, Wang J, Sun HC, Paschon DE, Guschin DY, Gregory PD, Kotton DN, Holmes MC, Sorscher EJ, Davis BR. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Reports. 2015;4(4):569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E, Dargitz CT, Wright R, Khanna A, Gage FH, Verma IM. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep. 2015;12(9):1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordonez A, Hannan NR, Rouhani FJ, Darche S, Alexander G, Marciniak SJ, Fusaki N, Hasegawa M, Holmes MC, Di Santo JP, Lomas DA, Bradley A, Vallier L. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478(7369):391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen JR, Tang ZH, Zheng J, Shi HS, Ding J, Qian XD, Zhang C, Chen JL, Wang CC, Li L, Chen JZ, Yin SK, Shao JZ, Huang TS, Chen P, Guan MX, Wang JF. Effects of genetic correction on the differentiation of hair cell-like cells from iPSCs with myo15a mutation. Cell Death Differ. 2016;23(8):1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kotini AG, Chang CJ, Boussaad I, Delrow JJ, Dolezal EK, Nagulapally AB, Perna F, Fishbein GA, Klimek VM, Hawkins RD, Huangfu D, Murry CE, Graubert T, Nimer SD, Papapetrou EP. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol. 2015;33(6):646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sebastiano V, Zhen HH, Haddad B, Bashkirova E, Melo SP, Wang P, Leung TL, Siprashvili Z, Tichy A, Li J, Ameen M, Hawkins J, Lee S, Li L, Schwertschkow A, Bauer G, Lisowski L, Kay MA, Kim SK, Lane AT, Wernig M, Oro AE. Human col7a1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, Pearl JR, Rebar EJ, Byron M, Gregory PD, Brown CJ, Urnov FD, Hall LL, Lawrence JB. Translating dosage compensation to trisomy 21. Nature. 2013;500(7462):296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, Kohn DB, Nakano A, Nelson SF, Miceli MC, Spencer MJ, Pyle AD. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hIPSc- derived muscle cells. Cell Stem Cell. 2016;18(4):533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS. Functional correction of large factor viii gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17(2):213–220. [DOI] [PubMed] [Google Scholar]
- 100. Chen C, Jiang P, Xue H, Peterson SE, Tran HT, McCann AE, Parast MM, Li S, Pleasure DE, Laurent LC, Loring JF, Liu Y, Deng W. Role of astroglia in Down syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat Commun. 2014;5:4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19(2):258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. [DOI] [PubMed] [Google Scholar]
- 104. Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15(2):215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10(10):973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12(12):1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of leopard syndrome. Nature. 2010;465(7299):808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, Goldstein LS. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, Takahashi K, Asaka I, Aoi T, Watanabe A, Watanabe K, Kadoya C, Nakano R, Watanabe D, Maruyama K, Hori O, Hibino S, Choshi T, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular abeta and differential drug responsiveness. Cell Stem Cell. 2013;12(4):487–496. [DOI] [PubMed] [Google Scholar]
- 112. Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkela E, Hyttinen J, Kontula K, Swan H, Conklin BR, Yamanaka S, Silvennoinen O, Aalto-Setala K. 2012. Model for long qt syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5(2):220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, Tabar V, Sadelain M, Studer L. Modelling pathogenesis and treatment of familial dysautonomia using patient-37 specific iPSCs. Nature. 2009;461(7262):402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-qt 6 syndrome. N Engl J Med. 2010;363(15):1397–1409. [DOI] [PubMed] [Google Scholar]
- 116. Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120(9):3127–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Shi Y, Kirwan P, Smith J, MacLean G, Orkin SH, Livesey FJ. A human stem cell model of early Alzheimer’s disease pathology in Down syndrome. Sci Transl Med. 2012;4(124):124ra129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4(130):130ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Weick JP, Held DL, Bonadurer GF, 3 rd, Doers ME, Liu Y, Maguire C, Clark A, Knackert JA, Molinarolo K, Musser M, Yao L, Yin Y, Lu J, Zhang X, Zhang SC, Bhattacharyya A. Deficits in human trisomy 21 iPSCs and neurons. Proc Natl Acad Sci USA. 2013;110(24):9962–9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, Makri G, Nauen D, Yu H, Guzman E, Chiang CH, Yoritomo N, Kaibuchi K, Zou J, Christian KM, Cheng L, Ross CA, Margolis RL, et al. 2014. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515(7527):414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yahata N, Asai M, Kitaoka S, Takahashi K, Asaka I, Hioki H, Kaneko T, Maruyama K, Saido TC, Nakahata T, Asada T, Yamanaka S, Iwata N, Inoue H. Anti-abeta drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer’s disease. PLoS One. 2011;6(9):e25788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6(4):896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, Zhang L, Guschin D, Fong LK, Vu BJ, Meng X, Urnov FD, Rebar EJ, Gregory PD, Zhang HS, Jaenisch R. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146(2):318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR, 3rd, Nakanishi N, Andreyev AY, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155(6):1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, Hoing S, Hargus G, Heck SA, Dhingra A, Wu G, Muller S, Brockmann K, Kluba T, Maisel M, Kruger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki OE, Klingauf J, Kuhlmann T, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12(3):354–367. [DOI] [PubMed] [Google Scholar]
- 127. Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, Hu S, Kay MA, Urnov FD, Shinnawi R, Gold JD, Gepstein L, Wu JC. Genome editing of isogenic human induced pluripotent stem cells recapitulates long qt phenotype for drug testing. J Am Coll Cardiol. 2014;64(5):451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Themeli M, Kloss CC, Ciriello G, Fedorov VD, Perna F, Gonen M, Sadelain M. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol. 2013;31(10):928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hotta A, Yamanaka S. From genomics to gene therapy: Induced pluripotent stem cells meet genome editing. Annu Rev Genet. 2015;49:47–70. [DOI] [PubMed] [Google Scholar]
- 132. Doi D, Samata B, Katsukawa M, Kikuchi T, Morizane A, Ono Y, Sekiguchi K, Nakagawa M, Parmar M, Takahashi J. Isolation of human induced pluripotent stem cell-derived dopaminergic progenitors by cell sorting for successful transplantation. Stem Cell Reports. 2014;2(3):337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Samata B, Doi D, Nishimura K, Kikuchi T, Watanabe A, Sakamoto Y, Kakuta J, Ono Y, Takahashi J. Purification of functional human ES and iPSC-derived midbrain dopaminergic progenitors using LRTM1. Nat Commun. 2016;7:13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Li C, Guan X, Du T, Jin W, Wu B, Liu Y, Wang P, Hu B, Griffin GE, Shattock RJ, Hu Q. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol. 2015;96(8):2381–2393. [DOI] [PubMed] [Google Scholar]
- 135. Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. Ccr5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9(12):e115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA. Sox17 is a critical specifier of human primordial 10 germ cell fate. Cell. 2015;160(1–2):253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]