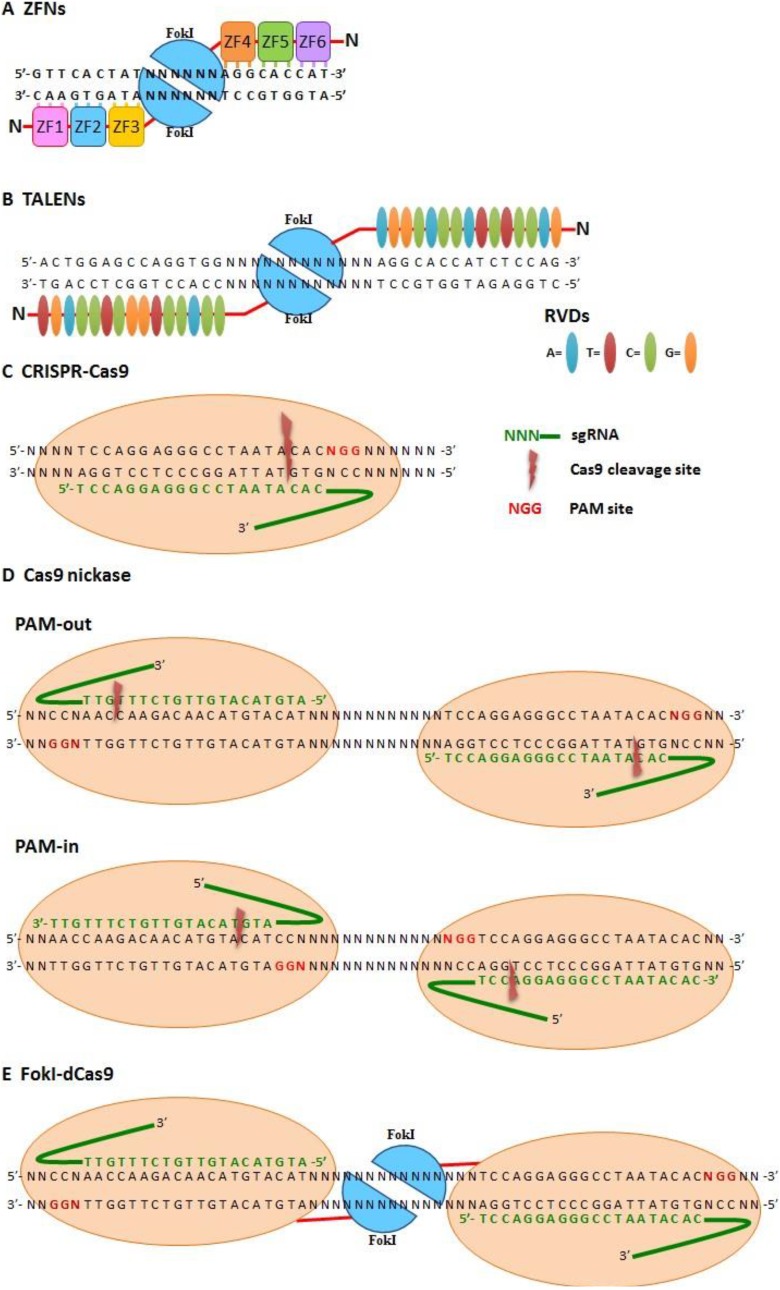
Fig. 2.
Site-specific nuclease (SSN)-based genome editing tools. (A) Zinc-finger nucleases (ZFNs). Two zinc-finger nucleases (ZFNs) cooperate for site-specific recognition with a 3-bp pairing/zinc finger and dimerized FokI to create double-strand breaks (DSBs) on DNA. (B) Transcription activator-like effector-based nucleases (TALENs). Two TALENs cooperate for site-specific recognition with 1-bp pairing/repeat variable di-residues (RVDs) and dimerized FokI to create DSBs on DNA. (C–E) Cas9-based SSN system. sgRNA recognizes specific sites via Watson–Crick pairing and cleavage target DNA (C) with wild-type Cas9 (which makes DSB), (D) Cas9 nickase (double-strand nicking with PAM-out and PAM-in orientations), and (E) FokI-dCas9 (which creates DSBs with dimerized FokI).