Figure 2.
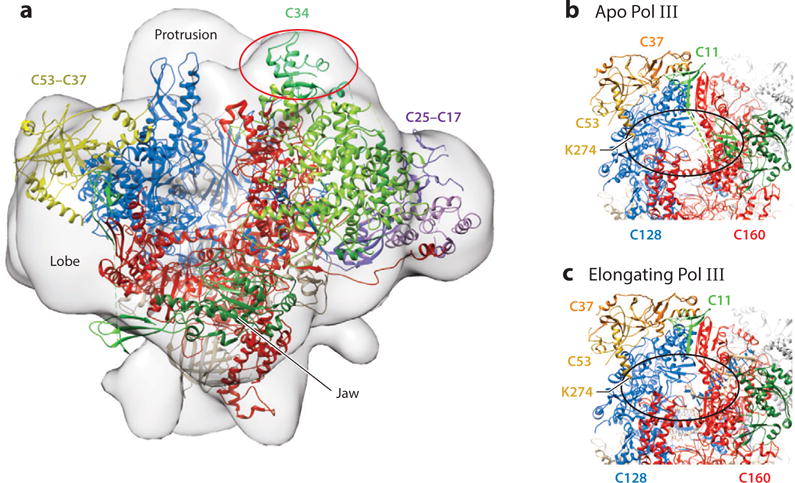
Structures of yeast Pol III. A Pol III-Maf1 complex. The 18.5Å resolution map (EMD-1755) is presented in the front view (15). The 4.7Å cryoEM structure of apo Pol III in the open form (17) was fitted into the envelope of the Pol III-Maf1 complex using Chimera. After fitting, 1806 out of 38434 atoms remained outside the contour. The DNA binding cleft comprising Rpc160 (red) and Rpc128 (blue) is viewed from the perspective of downstream DNA with the Wall in the rear. CryoEM difference density attributed to Maf1 (15) is shown in the oval (red) and lies behind the ribbon structure of Rpc34 (green) which is entirely outside the envelope in this fit. Rpc53/37 (yellow) and Rpc25/17 (purple) are shown for orientation. B and C. CryoEM structures of apo and elongating Pol III forms indicate mobility of the Rpc11 C-terminal zinc-binding domain. The Rpc53-Rpc37 subcomplex is positioned on the lobe of Rpc128. The two zinc ribbon domains of Rpc11 in the apo structure are connected by a linker (dashed line) which was not resolved. Density corresponding to the C-terminal domain of Rpc11 and the linker are missing in the elongating complex. Density for Rpc53 begins at K274 as shown. Photocrosslinking data place the Rpc53 phosphosites at amino acids 224, 228 and 232 within reach of Rpc160 and Rpc128, predicted to be within the oval. It is proposed that phosphorylation of these sites may affect the dynamics of the Rpc11 linker and its C-terminal zinc ribbon domain.
