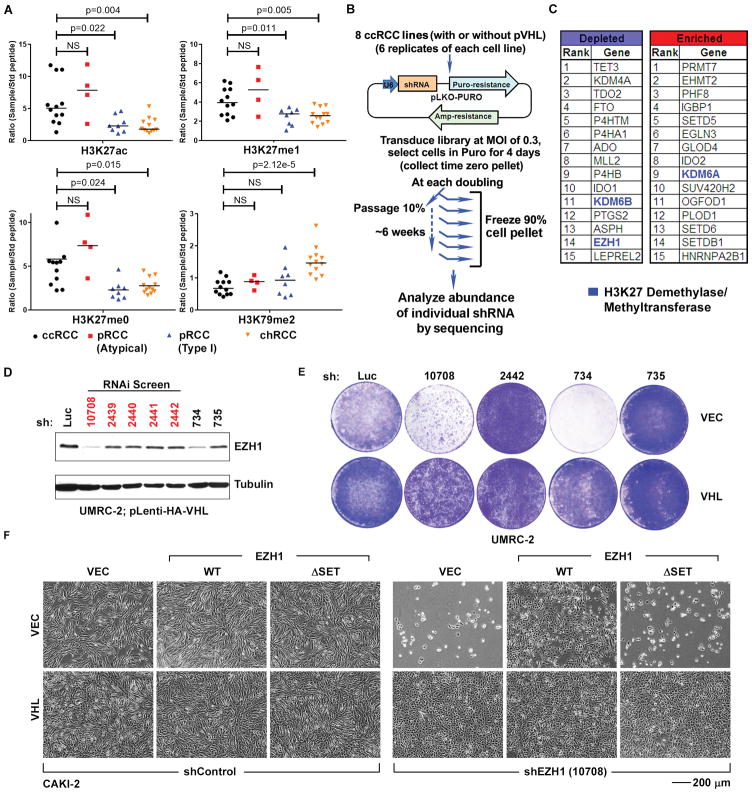
Fig. 1. VHL−/− ccRCC cells exhibit altered H3K27 modifications and increased dependence on EZH1.
(A) Quantification of the indicated histone modifications, as measured by mass spectrometric analysis (described in fig. S1), in renal tumor samples annotated as clear cell (ccRCC), typical papillary (pRCC, Type 1), atypical papillary (pRCC, atypical), and chromophobe (chRCC) renal carcinomas. p-values were calculated using Students t-test, and p<0.05 was considered significant. NS: Not Significant.
(B) shRNA screen schema.
(C) Genes whose shRNAs exhibited the greatest depletion or enrichment in the VHL−/− ccRCC relative to isogenic pVHL-proficient cells as determined by RIGER.
(D) Immunoblots of UMRC-2 cells that were lentivirally infected to produce pVHL and then stably infected to produce the indicated EZH1 shRNAs or an shRNA against firefly luciferase (Luc). The shRNAs in the screen are indicated in red.
(E) Crystal violet staining of UMRC-2 that were lentivirally infected to produce pVHL (VHL) or infected with an empty lentivirus (VEC) and then stably infected to produce the indicated EZH1 shRNAs or an shRNA against firefly luciferase (Luc).
(F) Photomicrographs of CAKI-2 cells that were infected to produce exogenous wild-type (WT) EZH1, catalytic-dead EZH1 (ΔSET), or with the empty vector (VEC) and then with a lentivirus encoding an EZH1 shRNA (10708) or control shRNA. The EZH1 WT vector contained synonymous mutations in the sequence recognized by the 10708 shRNA. The ΔSET mutation also eliminates the 10708 recognition sequence.